The landscape of human proteins interacting with viruses and other pathogens - PubMed (original) (raw)
The landscape of human proteins interacting with viruses and other pathogens
Matthew D Dyer et al. PLoS Pathog. 2008.
Abstract
Infectious diseases result in millions of deaths each year. Mechanisms of infection have been studied in detail for many pathogens. However, many questions are relatively unexplored. What are the properties of human proteins that interact with pathogens? Do pathogens interact with certain functional classes of human proteins? Which infection mechanisms and pathways are commonly triggered by multiple pathogens? In this paper, to our knowledge, we provide the first study of the landscape of human proteins interacting with pathogens. We integrate human-pathogen protein-protein interactions (PPIs) for 190 pathogen strains from seven public databases. Nearly all of the 10,477 human-pathogen PPIs are for viral systems (98.3%), with the majority belonging to the human-HIV system (77.9%). We find that both viral and bacterial pathogens tend to interact with hubs (proteins with many interacting partners) and bottlenecks (proteins that are central to many paths in the network) in the human PPI network. We construct separate sets of human proteins interacting with bacterial pathogens, viral pathogens, and those interacting with multiple bacteria and with multiple viruses. Gene Ontology functions enriched in these sets reveal a number of processes, such as cell cycle regulation, nuclear transport, and immune response that participate in interactions with different pathogens. Our results provide the first global view of strategies used by pathogens to subvert human cellular processes and infect human cells. Supplementary data accompanying this paper is available at http://staff.vbi.vt.edu/dyermd/publications/dyer2008a.html.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
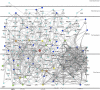
Figure 1. Human Proteins Interacting with Multiple Viral Pathogen Groups
The network of interactions between human proteins interacting with at least two viral pathogen groups. The size and color of a protein denote the number of pathogen groups that interact with it: light blue is two, dark blue is three, green is four, yellow is five, orange is six, and red is seven.
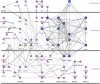
Figure 2. Human Proteins Interacting with Bacterial Pathogen Groups
The network of interactions between human proteins interacting with at least one bacterial pathogen group. The size and color of a protein denote the number of pathogen groups that interact with it: purple is one, light blue is two, dark blue is three, and green is four.
Figure 3. Degree and Centrality Distributions
Cumulative log-log distributions of (A) node degrees and (B) centralities for four subsets of nodes in the human PPI network: (i) red pluses are the set of all proteins in the network; (ii) green squares correspond to the viral set; (iii) blue crosses are for the bacterial set, and (iv) magenta squares are for the multiviral set. Numbers in parentheses represent the number of proteins in each set. The fraction of proteins at a particular value of degree or centrality is the number of proteins having that value or greater divided by the number of proteins in the set.
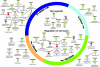
Figure 4. Human Cell Cycle Proteins Interacting with Multiple Viral Pathogen Groups
Enriched network of human proteins annotated with “cell cycle.” The subset of proteins labeled as “Non-specific” are those not annotated with any function more specific than “cell cycle” in GO. If a protein participates in multiple phases, then it appears in each phase. An edge connecting two proteins denotes a known interaction in the human PPI network. Human proteins highlighted in red are those known to be involved in the induction of apoptosis.
Figure 5. Human Nuclear Membrane Proteins Interacting with Multiple Viral Pathogen Groups
Enriched network of human proteins annotated with “nuclear transport” (blue), “nuclear membrane part” (green), “protein import” (orange), and “nuclear pore” (red). An edge connecting two proteins denotes a known interaction in the human PPI network.
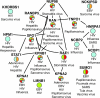
Figure 6. Human Immune System Proteins Interacting with Multiple Bacterial Pathogen Groups
Enriched network of human proteins annotated with “immune system process” (red), “response to wounding” (orange), “immune response” (green), and “I-κB kinase/NF-κB cascade” (blue). The proteins in the black box form a dense network of PPIs; we have left these out for clarity. An edge connecting two proteins denotes a known interaction in the human PPI network.
Similar articles
- Computational prediction of host-pathogen protein-protein interactions.
Dyer MD, Murali TM, Sobral BW. Dyer MD, et al. Bioinformatics. 2007 Jul 1;23(13):i159-66. doi: 10.1093/bioinformatics/btm208. Bioinformatics. 2007. PMID: 17646292 - Molecular principles of human virus protein-protein interactions.
Halehalli RR, Nagarajaram HA. Halehalli RR, et al. Bioinformatics. 2015 Apr 1;31(7):1025-33. doi: 10.1093/bioinformatics/btu763. Epub 2014 Nov 21. Bioinformatics. 2015. PMID: 25417202 - Host pathogen protein interactions predicted by comparative modeling.
Davis FP, Barkan DT, Eswar N, McKerrow JH, Sali A. Davis FP, et al. Protein Sci. 2007 Dec;16(12):2585-96. doi: 10.1110/ps.073228407. Epub 2007 Oct 26. Protein Sci. 2007. PMID: 17965183 Free PMC article. - Complement evasion by human pathogens.
Lambris JD, Ricklin D, Geisbrecht BV. Lambris JD, et al. Nat Rev Microbiol. 2008 Feb;6(2):132-42. doi: 10.1038/nrmicro1824. Nat Rev Microbiol. 2008. PMID: 18197169 Free PMC article. Review. - Current status and future perspectives of computational studies on human-virus protein-protein interactions.
Lian X, Yang X, Yang S, Zhang Z. Lian X, et al. Brief Bioinform. 2021 Sep 2;22(5):bbab029. doi: 10.1093/bib/bbab029. Brief Bioinform. 2021. PMID: 33693490 Review.
Cited by
- From Viral Infections to Alzheimer's Disease: Unveiling the Mechanistic Links Through Systems Bioinformatics.
Onisiforou A, Zanos P. Onisiforou A, et al. J Infect Dis. 2024 Sep 10;230(Supplement_2):S128-S140. doi: 10.1093/infdis/jiae242. J Infect Dis. 2024. PMID: 39255398 Free PMC article. - Adaptation of turnip mosaic virus to Arabidopsis thaliana involves rewiring of VPg-host proteome interactions.
Carrasco JL, Ambrós S, Gutiérrez PA, Elena SF. Carrasco JL, et al. Virus Evol. 2024 Jul 16;10(1):veae055. doi: 10.1093/ve/veae055. eCollection 2024. Virus Evol. 2024. PMID: 39091990 Free PMC article. - A microbial knowledge graph-based deep learning model for predicting candidate microbes for target hosts.
Pan J, Zhang Z, Li Y, Yu J, You Z, Li C, Wang S, Zhu M, Ren F, Zhang X, Sun Y, Wang S. Pan J, et al. Brief Bioinform. 2024 Mar 27;25(3):bbae119. doi: 10.1093/bib/bbae119. Brief Bioinform. 2024. PMID: 38555472 Free PMC article. - Exploring Viral-Host Protein Interactions as Antiviral Therapies: A Computational Perspective.
Idrees S, Chen H, Panth N, Paudel KR, Hansbro PM. Idrees S, et al. Microorganisms. 2024 Mar 21;12(3):630. doi: 10.3390/microorganisms12030630. Microorganisms. 2024. PMID: 38543681 Free PMC article. Review. - Prediction of motif-mediated viral mimicry through the integration of host-pathogen interactions.
Idrees S, Paudel KR, Hansbro PM. Idrees S, et al. Arch Microbiol. 2024 Feb 9;206(3):94. doi: 10.1007/s00203-024-03832-9. Arch Microbiol. 2024. PMID: 38334822 Free PMC article.
References
- LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, et al. A protein interaction network of the malaria parasite Plasmodium falciparum . Nature. 2005;438:103–107. - PubMed
- Filippova M, Parkhurst L, Duerksen-Hughes PJ. The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J Biol Chem. 2004;279:25729–25744. - PubMed
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, et al. The innate immune response to bacterial flagellin is mediated by toll-like receptor 5. Nature. 2001;410:1099–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources