The effect of inappropriate calibration: three case studies in molecular ecology - PubMed (original) (raw)
The effect of inappropriate calibration: three case studies in molecular ecology
Simon Y W Ho et al. PLoS One. 2008.
Abstract
Time-scales estimated from sequence data play an important role in molecular ecology. They can be used to draw correlations between evolutionary and palaeoclimatic events, to measure the tempo of speciation, and to study the demographic history of an endangered species. In all of these studies, it is paramount to have accurate estimates of time-scales and substitution rates. Molecular ecological studies typically focus on intraspecific data that have evolved on genealogical scales, but often these studies inappropriately employ deep fossil calibrations or canonical substitution rates (e.g., 1% per million years for birds and mammals) for calibrating estimates of divergence times. These approaches can yield misleading estimates of molecular time-scales, with significant impacts on subsequent evolutionary and ecological inferences. We illustrate this calibration problem using three case studies: avian speciation in the late Pleistocene, the demographic history of bowhead whales, and the Pleistocene biogeography of brown bears. For each data set, we compare the date estimates that are obtained using internal and external calibration points. In all three cases, the conclusions are significantly altered by the application of revised, internally-calibrated substitution rates. Collectively, the results emphasise the importance of judicious selection of calibrations for analyses of recent evolutionary events.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
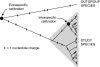
Figure 1. Phylogenetic tree illustrating the impact of using extraspecific and intraspecific calibration points.
The tree shows the locations of nucleotide changes (small vertical bars). The nucleotide changes within the study species represent segregating sites, some of which will be fixed in the long term, but most of which will be removed by drift or selection. The changes between the study species and outgroup species represent substitutions. If an estimate of the evolutionary rate is calibrated using an external calibration point, such as the split between the study and outgroup species, then the intraspecific rate will be underestimated. This will lead to an overestimation of times to divergence, including the age of the most recent common ancestor of the study species.
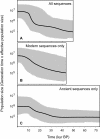
Figure 2. Bayesian skyline plots showing the recent demographic history of bowhead whales, estimated using phylogenetic analysis of three alignments of the mitochondrial control region: (a) combined alignment of 68 modern haplotypes and 99 radiocarbon-dated, ancient DNA sequences; (b) modern sequences only; and (c) ancient sequences only.
All three plots are drawn to the vertical and horizontal scales.
Figure 3. Maximum clade credibility tree from Bayesian analysis of mitochondrial control region sequences from 56 modern and 51 ancient brown bear samples, with a time scale calibrated using the radiocarbon dates of the ancient sequences.
Major clades and their geographic localities are given. Posterior probabilities are given for the nodes A-N, which are referred to in the text and in Table 2.
Similar articles
- Evaluating fossil calibrations for dating phylogenies in light of rates of molecular evolution: a comparison of three approaches.
Lukoschek V, Scott Keogh J, Avise JC. Lukoschek V, et al. Syst Biol. 2012 Jan;61(1):22-43. doi: 10.1093/sysbio/syr075. Epub 2011 Aug 13. Syst Biol. 2012. PMID: 21840843 - The fossilized birth-death process for coherent calibration of divergence-time estimates.
Heath TA, Huelsenbeck JP, Stadler T. Heath TA, et al. Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):E2957-66. doi: 10.1073/pnas.1319091111. Epub 2014 Jul 9. Proc Natl Acad Sci U S A. 2014. PMID: 25009181 Free PMC article. - Assessing the quality of molecular divergence time estimates by fossil calibrations and fossil-based model selection.
Near TJ, Sanderson MJ. Near TJ, et al. Philos Trans R Soc Lond B Biol Sci. 2004 Oct 29;359(1450):1477-83. doi: 10.1098/rstb.2004.1523. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15519966 Free PMC article. - The early Eocene birds of the Messel fossil site: a 48 million-year-old bird community adds a temporal perspective to the evolution of tropical avifaunas.
Mayr G. Mayr G. Biol Rev Camb Philos Soc. 2017 May;92(2):1174-1188. doi: 10.1111/brv.12274. Epub 2016 Apr 8. Biol Rev Camb Philos Soc. 2017. PMID: 27062331 Review. - Inferences from tip-calibrated phylogenies: a review and a practical guide.
Rieux A, Balloux F. Rieux A, et al. Mol Ecol. 2016 May;25(9):1911-24. doi: 10.1111/mec.13586. Epub 2016 Apr 20. Mol Ecol. 2016. PMID: 26880113 Free PMC article. Review.
Cited by
- Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses.
Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, Rambaut A. Firth C, et al. Mol Biol Evol. 2010 Sep;27(9):2038-51. doi: 10.1093/molbev/msq088. Epub 2010 Apr 2. Mol Biol Evol. 2010. PMID: 20363828 Free PMC article. - Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species.
Horn S, Durka W, Wolf R, Ermala A, Stubbe A, Stubbe M, Hofreiter M. Horn S, et al. PLoS One. 2011 Jan 28;6(1):e14622. doi: 10.1371/journal.pone.0014622. PLoS One. 2011. PMID: 21307956 Free PMC article. - Diabolical survival in Death Valley: recent pupfish colonization, gene flow and genetic assimilation in the smallest species range on earth.
Martin CH, Crawford JE, Turner BJ, Simons LH. Martin CH, et al. Proc Biol Sci. 2016 Jan 27;283(1823):20152334. doi: 10.1098/rspb.2015.2334. Proc Biol Sci. 2016. PMID: 26817777 Free PMC article. - Sexual species are separated by larger genetic gaps than asexual species in rotifers.
Tang CQ, Obertegger U, Fontaneto D, Barraclough TG. Tang CQ, et al. Evolution. 2014 Oct;68(10):2901-16. doi: 10.1111/evo.12483. Epub 2014 Jul 25. Evolution. 2014. PMID: 24975991 Free PMC article.
References
- Lovette IJ. Glacial cycles and the tempo of avian speciation. Trends Ecol Evol. 2005;20:57–59. - PubMed
- Hofreiter M, Rabeder G, Jaenicke-Despres V, Withalm G, Nagel D, et al. Evidence for reproductive isolation between cave bear populations. Curr Biol. 2004;14:40–43. - PubMed
- Saarma U, Ho SYW, Pybus OG, Kaljuste M, Tumanov IM, et al. Mitogenetic structure of brown bears (Ursus arctos L.) in north-eastern Europe and a new time-frame for the formation of brown bear lineages. Mol Ecol. 2007;16:401–413. - PubMed
- Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. - PubMed
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources