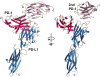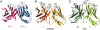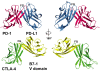The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors - PubMed (original) (raw)
The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors
David Yin-Wei Lin et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Signaling through the programmed death 1 (PD-1) inhibitory receptor upon binding its ligand, PD-L1, suppresses immune responses against autoantigens and tumors and plays an important role in the maintenance of peripheral immune tolerance. Release from PD-1 inhibitory signaling revives "exhausted" virus-specific T cells in chronic viral infections. Here we present the crystal structure of murine PD-1 in complex with human PD-L1. PD-1 and PD-L1 interact through the conserved front and side of their Ig variable (IgV) domains, as do the IgV domains of antibodies and T cell receptors. This places the loops at the ends of the IgV domains on the same side of the PD-1/PD-L1 complex, forming a surface that is similar to the antigen-binding surface of antibodies and T cell receptors. Mapping conserved residues allowed the identification of residues that are important in forming the PD-1/PD-L1 interface. Based on the structure, we show that some reported loss-of-binding mutations involve the PD-1/PD-L1 interaction but that others compromise protein folding. The PD-1/PD-L1 interaction described here may be blocked by antibodies or by designed small-molecule drugs to lower inhibitory signaling that results in a stronger immune response. The immune receptor-like loops offer a new surface for further study and potentially the design of molecules that would affect PD-1/PD-L1 complex formation and thereby modulate the immune response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Two views of the PD-1/PD-L1 complex. The two single-domain PD-1 molecules in the asymmetric unit of the crystal are shown in red (PD-1) and violet (second PD-1). The two-domain PD-L1 molecule is shown in blue. The strands of the two β-sheets of PD-1 are labeled ABED and A′GFCC′C″. The strands of the two β-sheets of the PD-L1 V domain are labeled AGFCC′C″ and BED. Note the tenuous contacts that the second PD-1 makes to PD-1 and PD-L1, seen best on the right side of the figure. N-linked glycosylation sites (gold) are at PD-1 residues 49, 58, 74, and 116 and at PD-L1 residue 35. Carbohydrate at any of the five potential sites is predicted not to interfere with the formation of the complex. The view at right is after a rotation of 45° around the vertical axis. The N and C termini are identified.
Fig. 2.
The PD-1/PD-L1 interface. Shown is a stereoview of the PD-1/PD-L1 interface showing side chains of residues on β-strands (CC′FG) of PD-1 (red) and on β-strands (GFCC′, left to right) of PD-L1 (blue) that make contacts. Interacting PD-1 side chains (pink) and PD-L1 (light blue) are shown; for clarity a few side chains are not shown. Dotted lines (yellow) indicate hydrogen bonds formed in the interface and with a water molecule.
Fig. 3.
The IgV domains of PD1/PD-L1 are similar to antigen receptors. (A) The loops at the ends of the PD-1 domain and the first domain of PD-L1 are located on the same side of the complex. (B) For comparison, the Fv or VHVL portion of a representative antibody [PDB ID code 1G7J (31)] found in the search of the database was superimposed on the Fv-like portion of the PD-1/PD-L1 complex to yield an rms difference of 2.6 Å over 180 pairs of α-carbon pairs. (C) The VαVβ portion of a representative TCR [PDB ID code 2BNU (32)] was superimposed on the Fv-like part of the PD-1/PD-L1 complex with an rms difference of 3.1 Å over 191 Cα pairs. In B and C, after superimposing on to PD-1/PD-L1, the models were translated apart for viewing. The loops that are antigen receptor CDRs are labeled (CDR1, CDR2, and CDR3). The search of the database was performed with the PD-1/PD-L1 Fv-like domain as query and all of the targets considered as two-domain rigid bodies.
Fig. 4.
A comparison of the PD-1/PD-L1 and CTLA-4/B7-1 variable-like (V) domain interactions. (Left) the two complexes were overlaid by superimposing PD-1 (red) on to CTLA-4 (green), then translated apart vertically for viewing. (Right) The two complexes were rotated by 180°. Note the marked difference in the location of the B7-1 V domain (yellow) in comparison to the V domain of PD-L1 (blue).
Similar articles
- Analysis of the role of negative T cell costimulatory pathways in CD4 and CD8 T cell-mediated alloimmune responses in vivo.
Ito T, Ueno T, Clarkson MR, Yuan X, Jurewicz MM, Yagita H, Azuma M, Sharpe AH, Auchincloss H Jr, Sayegh MH, Najafian N. Ito T, et al. J Immunol. 2005 Jun 1;174(11):6648-56. doi: 10.4049/jimmunol.174.11.6648. J Immunol. 2005. PMID: 15905503 - Anti-tumor immunotherapy by blockade of the PD-1/PD-L1 pathway with recombinant human PD-1-IgV.
Zhang C, Wu S, Xue X, Li M, Qin X, Li W, Han W, Zhang Y. Zhang C, et al. Cytotherapy. 2008;10(7):711-9. doi: 10.1080/14653240802320237. Cytotherapy. 2008. PMID: 18841514 - PD-L1 and PD-L2 differ in their molecular mechanisms of interaction with PD-1.
Ghiotto M, Gauthier L, Serriari N, Pastor S, Truneh A, Nunès JA, Olive D. Ghiotto M, et al. Int Immunol. 2010 Aug;22(8):651-60. doi: 10.1093/intimm/dxq049. Epub 2010 Jun 29. Int Immunol. 2010. PMID: 20587542 Free PMC article. - Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways.
Fife BT, Bluestone JA. Fife BT, et al. Immunol Rev. 2008 Aug;224:166-82. doi: 10.1111/j.1600-065X.2008.00662.x. Immunol Rev. 2008. PMID: 18759926 Review. - Subverting the B7-H1/PD-1 pathway in advanced melanoma and kidney cancer.
Harshman LC, Choueiri TK, Drake C, Stephen Hodi F Jr. Harshman LC, et al. Cancer J. 2014 Jul-Aug;20(4):272-80. doi: 10.1097/PPO.0000000000000055. Cancer J. 2014. PMID: 25098288 Free PMC article. Review.
Cited by
- Network Meta-analysis Comparing Efficacy, Safety and Tolerability of Anti-PD-1/PD-L1 Antibodies in Solid Cancers.
Al-Showbaki L, Nadler MB, Desnoyers A, Almugbel FA, Cescon DW, Amir E. Al-Showbaki L, et al. J Cancer. 2021 May 19;12(14):4372-4378. doi: 10.7150/jca.57413. eCollection 2021. J Cancer. 2021. PMID: 34093837 Free PMC article. - PD-1 derived CA-170 is an oral immune checkpoint inhibitor that exhibits preclinical anti-tumor efficacy.
Sasikumar PG, Sudarshan NS, Adurthi S, Ramachandra RK, Samiulla DS, Lakshminarasimhan A, Ramanathan A, Chandrasekhar T, Dhudashiya AA, Talapati SR, Gowda N, Palakolanu S, Mani J, Srinivasrao B, Joseph D, Kumar N, Nair R, Atreya HS, Gowda N, Ramachandra M. Sasikumar PG, et al. Commun Biol. 2021 Jun 8;4(1):699. doi: 10.1038/s42003-021-02191-1. Commun Biol. 2021. PMID: 34103659 Free PMC article. - PD-1 expression on mouse intratumoral NK cells and its effects on NK cell phenotype.
Wagner AK, Kadri N, Tibbitt C, van de Ven K, Bagawath-Singh S, Oliinyk D, LeGresley E, Campbell N, Trittel S, Riese P, Ribacke U, Sandalova T, Achour A, Kärre K, Chambers BJ. Wagner AK, et al. iScience. 2022 Sep 16;25(10):105137. doi: 10.1016/j.isci.2022.105137. eCollection 2022 Oct 21. iScience. 2022. PMID: 36185379 Free PMC article. - Immune Checkpoint PD-1/PD-L1 CTLA-4/CD80 are Blocked by Rhus verniciflua Stokes and its Active Compounds.
Li W, Kim TI, Kim JH, Chung HS. Li W, et al. Molecules. 2019 Nov 9;24(22):4062. doi: 10.3390/molecules24224062. Molecules. 2019. PMID: 31717574 Free PMC article. - B7 family protein glycosylation: Promising novel targets in tumor treatment.
Xiao L, Guan X, Xiang M, Wang Q, Long Q, Yue C, Chen L, Liu J, Liao C. Xiao L, et al. Front Immunol. 2022 Dec 6;13:1088560. doi: 10.3389/fimmu.2022.1088560. eCollection 2022. Front Immunol. 2022. PMID: 36561746 Free PMC article. Review.
References
- Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. - PubMed
- Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. - PubMed
- Nishimura H, et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials