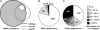PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis - PubMed (original) (raw)
Comparative Study
. 2008 Feb 26;105(8):3145-50.
doi: 10.1073/pnas.0709632105. Epub 2008 Feb 19.
Affiliations
- PMID: 18287047
- PMCID: PMC2268599
- DOI: 10.1073/pnas.0709632105
Comparative Study
PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis
Rebecca A Mosher et al. Proc Natl Acad Sci U S A. 2008.
Abstract
DNA-dependent RNA polymerase (Pol)IV in Arabidopsis exists in two isoforms (PolIVa and PolIVb), with NRPD1a and NRPD1b as their respective largest subunits. Both isoforms are implicated in production and activity of siRNAs and in RNA-directed DNA methylation (RdDM). Deep sequence analysis of siRNAs in WT Arabidopsis flowers and in nrpd1a and nrpd1b mutants identified >4,200 loci producing siRNAs in a PolIV-dependent manner, with PolIVb reinforcing siRNA production by PolIVa. Transposable element identity and pericentromeric localization are both features that predispose a locus for siRNA production via PolIV proteins and determine the extent to which siRNA production relies on PolIVb. Detailed analysis of DNA methylation at PolIV-dependent loci revealed unexpected deviations from the previously noted association of PolIVb-dependent siRNA production and RdDM. Notably, PolIVb functions independently in DNA methylation and siRNA generation. Additionally, we have uncovered siRNA-directed loss of DNA methylation, a process requiring both PolIV isoforms. From these findings, we infer that the role of PolIVb in siRNA production is secondary to a role in chromatin modification and is influenced by chromatin context.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
PolIVa is required for siRNA accumulation at most endogenous loci. (A) Representative Venn diagram of siRNA-generating loci. Solid line, loci with siRNAs in WT; dotted line, loci with siRNAs at WT or a greater level in nrpd1a; dashed line, loci with siRNAs present at WT or greater level in nrpd1b. (B and C) Pie charts depicting level of siRNA representation at loci requiring at least one PolIV isoform (gray region in A).
Fig. 2.
Genetic dependence of PolIV-dependent loci. siRNA Northern blots of mixed floral tissue confirm that all tested PolIV-dependent loci lose siRNAs in nrpd1a. They also require NRPD2, RDR2, and DCL3 for siRNA accumulation but show various degrees of dependence on NRPD1b. The size markers are estimates based on 20- and 30-nt RNA oligos.
Fig. 3.
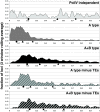
PolIV-dependent loci exhibit differential association with transposable elements but no inherent repetitive character. (A) Over- or underrepresentation of genomic features at PolIV-independent, A type, and A+B type loci. The locus positions were randomized 100 times, and the associated genomic elements were counted to calculate Z scores: Z = (observed − averagerand)/SDrand. (B and C) Histograms of loci with respect to their genomic repetitiveness. Each locus was BLASTed against the Arabidopsis genome, and the resulting matches (E < 0.001) were counted and plotted.
Fig. 4.
PolIV-dependent loci are pericentromerically localized independent of transposable element character. Histograms of relative centromeric bias of sRNA-generating loci. 0, centromere; 1, telomere. The rolling average of the fraction of loci in three 1% windows is plotted. The short arms of chromosomes 2 and 4 were removed for clarity (33). Circles, triangles, and stars mark the first, second, and third quartiles, respectively.
Fig. 5.
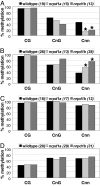
RNA-directed DNA methylation at PolIVa-dependent loci. Bisulfite sequencing of PolIV-dependent loci 00687 (A), 08002 (B), 10102 (C), and 04138 (D). Data from two biological replicates were combined. Asterisks mark changes that were statistically significant (two-tailed t test, P < 0.01) in both replicates. The total number of clones analyzed is in parenthesis.
Fig. 6.
Model of PolIV dependency of siRNA-generating loci. PolIV-independent epigenetic marks (blue stars) recruit PolIVa (pink ovals) to generate siRNAs. In the presence of PolIVb, additional epigenetic marks (yellow triangles) can reinforce recruitment of PolIVa. Additionally, RDR2-mediated amplification of siRNAs can occur when direct repeats are present in the transcript, as in LTR retroelement RNAs.
Similar articles
- PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit.
Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. Lahmy S, et al. Proc Natl Acad Sci U S A. 2009 Jan 20;106(3):941-6. doi: 10.1073/pnas.0810310106. Epub 2009 Jan 13. Proc Natl Acad Sci U S A. 2009. PMID: 19141635 Free PMC article. - NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis.
Eamens A, Vaistij FE, Jones L. Eamens A, et al. Plant J. 2008 Aug;55(4):596-606. doi: 10.1111/j.1365-313X.2008.03525.x. Epub 2008 Apr 22. Plant J. 2008. PMID: 18433438 - NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - Plant-specific multisubunit RNA polymerase in gene silencing.
Lahmy S, Bies-Etheve N, Lagrange T. Lahmy S, et al. Epigenetics. 2010 Jan 1;5(1):4-8. doi: 10.4161/epi.5.1.10435. Epub 2010 Jan 26. Epigenetics. 2010. PMID: 20173420 Review. - Non-coding RNA transcription and RNA-directed DNA methylation in Arabidopsis.
He XJ, Ma ZY, Liu ZW. He XJ, et al. Mol Plant. 2014 Sep;7(9):1406-1414. doi: 10.1093/mp/ssu075. Epub 2014 Jun 25. Mol Plant. 2014. PMID: 24966349 Review.
Cited by
- An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II.
Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. Huang L, et al. Nat Struct Mol Biol. 2009 Jan;16(1):91-3. doi: 10.1038/nsmb.1539. Epub 2008 Dec 14. Nat Struct Mol Biol. 2009. PMID: 19079263 - Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing.
Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Rowley MJ, et al. PLoS Genet. 2011 Jun;7(6):e1002120. doi: 10.1371/journal.pgen.1002120. Epub 2011 Jun 9. PLoS Genet. 2011. PMID: 21738482 Free PMC article. - RNAi in Plants: An Argonaute-Centered View.
Fang X, Qi Y. Fang X, et al. Plant Cell. 2016 Feb;28(2):272-85. doi: 10.1105/tpc.15.00920. Epub 2016 Feb 11. Plant Cell. 2016. PMID: 26869699 Free PMC article. Review. - An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein.
He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. He XJ, et al. Cell. 2009 May 1;137(3):498-508. doi: 10.1016/j.cell.2009.04.028. Cell. 2009. PMID: 19410546 Free PMC article. - The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis.
Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, He XJ. Zhang CJ, et al. EMBO J. 2013 Apr 17;32(8):1128-40. doi: 10.1038/emboj.2013.49. Epub 2013 Mar 22. EMBO J. 2013. PMID: 23524848 Free PMC article.
References
- Herr AJ. FEBS Lett. 2005;579:5879–5888. - PubMed
- Herr AJ, Jensen MB, Dalmay T, Baulcombe D. Science. 2005;308:118–120. - PubMed
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Nat Genet. 2005;37:761–765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases