Protein area occupancy at the center of the red blood cell membrane - PubMed (original) (raw)
Comparative Study
. 2008 Feb 26;105(8):2848-52.
doi: 10.1073/pnas.0712379105. Epub 2008 Feb 19.
Affiliations
- PMID: 18287056
- PMCID: PMC2268548
- DOI: 10.1073/pnas.0712379105
Comparative Study
Protein area occupancy at the center of the red blood cell membrane
Allison D Dupuy et al. Proc Natl Acad Sci U S A. 2008.
Abstract
In the Fluid Mosaic Model for biological membrane structure, proposed by Singer and Nicolson in 1972, the lipid bilayer is represented as a neutral two-dimensional solvent in which the proteins of the membrane are dispersed and distributed randomly. The model portrays the membrane as dominated by a membrane lipid bilayer, directly exposed to the aqueous environment, and only occasionally interrupted by transmembrane proteins. This view is reproduced in virtually every textbook in biochemistry and cell biology, yet some critical features have yet to be closely examined, including the key parameter of the relative occupancy of protein and lipid at the center of a natural membrane. Here we show that the area occupied by protein and lipid at the center of the human red blood cell (RBC) plasma membrane is at least approximately 23% protein and less than approximately 77% lipid. This measurement is in close agreement with previous estimates for the RBC plasma membrane and the recently published measurements for the synaptic vesicle. Given that transmembrane proteins are surrounded by phospholipids that are perturbed by their presence, the occupancy by protein of more than approximately 20% of the RBC plasma membrane and the synaptic vesicle plasma membrane implies that natural membrane bilayers may be more rigid and less fluid than has been thought for the past several decades, and that studies of pure lipid bilayers do not fully reveal the properties of lipids in membranes. Thus, it appears to be the case that membranes may be more mosaic than fluid, with little unperturbed phospholipid bilayer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
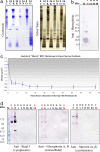
Fig. 1.
Time progression of membrane shaving. (a) Membrane washes. RBC open ghost membranes were washed in 1 M KCl (K), 10 mM NaOH (N), or 100 mM Na2CO3 (O), and then shaved with 3.0 mg/ml proteinase K in the respective wash buffer for 24 h (C, control, membranes were not washed). The membrane samples were then run on 1D SDS/PAGE gels and subjected to standard Coomassie and silver staining procedures. 0, no shaving with proteinase K; 24, membranes were shaved with 3.0 mg/ml proteinase K for 24 h. (b) Elimination of contamination from hemoglobin. RBC open ghost membranes washed (w) in 10 mM NaOH were shaved with 3.0 mg/ml proteinase K in 10 mM NaOH for various lengths of time up to 48 h. The membrane samples were then run on 1D SDS/PAGE gels and subjected to standard immunoblot analysis by using anti-hemoglobin (Sigma, H4890). Lane M, markers; lane H, hemoglobin supernatant (positive control for hemoglobin). (c) Time progression of membrane shaving by using buoyant density measurements from equilibrium centrifugation in linear sucrose gradients. RBC plasma membrane open ghosts were progressively shaved by proteolytic digestion with 3.0 mg/ml proteinase K. The proteolytic digestion was stopped with 5 mM PMSF as a function of time. At each time point, buoyant density measurements were made by using equilibrium centrifugation in continuous, linear sucrose gradients. Membranes were loaded on 5–45% continuous, linear sucrose gradients made by using the BioComp Gradient Master and spun at 215,578 × g for 24 h. Gradients were then fractionated by using the BioComp Gradient Fractionator. The density of sucrose at which the sample band of membranes was found by refractometry (equal to a buoyant density measurement for the membrane itself) was measured by using a temperature-controlled, small-volume, automatic refractometer (Rudolph Research Analytical, J57). The density at each time point was measured independently a total of ten times. (d) Time progression of membrane shaving by using immunoblot analysis. RBC open ghost membranes washed (w) in 10 mM NaOH and unwashed (u) RBC open ghost membranes were shaved with 3.0 mg/ml proteinase K in 10 mM NaOH for various lengths of time up to 24 h. 0, control, no proteinase K; s, shaved for 5 s. The membrane samples were then run on 1D SDS/PAGE gels and subjected to standard immunoblot analysis. Three different antibodies were analyzed in the immunoblot analysis including anti-band 3 (Sigma B9277) (cytoplasmic), anti-glycophorins A and B (Sigma G7650) (extracellular), and anti-spectrin (α and β) (Sigma S3396) (cytoskeleton). Lane M, markers.
Similar articles
- Is a fluid-mosaic model of biological membranes fully relevant? Studies on lipid organization in model and biological membranes.
Wiśniewska A, Draus J, Subczynski WK. Wiśniewska A, et al. Cell Mol Biol Lett. 2003;8(1):147-59. Cell Mol Biol Lett. 2003. PMID: 12655369 - Elastic deformation and failure of lipid bilayer membranes containing cholesterol.
Needham D, Nunn RS. Needham D, et al. Biophys J. 1990 Oct;58(4):997-1009. doi: 10.1016/S0006-3495(90)82444-9. Biophys J. 1990. PMID: 2249000 Free PMC article. - Molecular cloning of human plasma membrane phospholipid scramblase. A protein mediating transbilayer movement of plasma membrane phospholipids.
Zhou Q, Zhao J, Stout JG, Luhm RA, Wiedmer T, Sims PJ. Zhou Q, et al. J Biol Chem. 1997 Jul 18;272(29):18240-4. doi: 10.1074/jbc.272.29.18240. J Biol Chem. 1997. PMID: 9218461 - The basic structure and dynamics of cell membranes: an update of the Singer-Nicolson model.
Goñi FM. Goñi FM. Biochim Biophys Acta. 2014 Jun;1838(6):1467-76. doi: 10.1016/j.bbamem.2014.01.006. Epub 2014 Jan 16. Biochim Biophys Acta. 2014. PMID: 24440423 Review. - Structure and functional properties of diacylglycerols in membranes.
Goñi FM, Alonso A. Goñi FM, et al. Prog Lipid Res. 1999 Jan;38(1):1-48. doi: 10.1016/s0163-7827(98)00021-6. Prog Lipid Res. 1999. PMID: 10396601 Review.
Cited by
- The energetics of protein-lipid interactions as viewed by molecular simulations.
Corey RA, Stansfeld PJ, Sansom MSP. Corey RA, et al. Biochem Soc Trans. 2020 Feb 28;48(1):25-37. doi: 10.1042/BST20190149. Biochem Soc Trans. 2020. PMID: 31872229 Free PMC article. Review. - Massive formation of intracellular membrane vesicles in Escherichia coli by a monotopic membrane-bound lipid glycosyltransferase.
Eriksson HM, Wessman P, Ge C, Edwards K, Wieslander A. Eriksson HM, et al. J Biol Chem. 2009 Dec 4;284(49):33904-14. doi: 10.1074/jbc.M109.021618. Epub 2009 Sep 18. J Biol Chem. 2009. PMID: 19767390 Free PMC article. - Phosphatidylserine Asymmetry Promotes the Membrane Insertion of a Transmembrane Helix.
Scott HL, Heberle FA, Katsaras J, Barrera FN. Scott HL, et al. Biophys J. 2019 Apr 23;116(8):1495-1506. doi: 10.1016/j.bpj.2019.03.003. Epub 2019 Mar 19. Biophys J. 2019. PMID: 30954213 Free PMC article. - Packing Density of the Amyloid Precursor Protein in the Cell Membrane.
de Coninck D, Schmidt TH, Schloetel JG, Lang T. de Coninck D, et al. Biophys J. 2018 Mar 13;114(5):1128-1141. doi: 10.1016/j.bpj.2018.01.009. Biophys J. 2018. PMID: 29539399 Free PMC article. - Dawn of a New Era for Membrane Protein Design.
Sowlati-Hashjin S, Gandhi A, Garton M. Sowlati-Hashjin S, et al. Biodes Res. 2022 Apr 15;2022:9791435. doi: 10.34133/2022/9791435. eCollection 2022. Biodes Res. 2022. PMID: 37850134 Free PMC article. Review.
References
- Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. - PubMed
- Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. - PubMed
- Henderson R. The structure of the purple membrane from Halobacterium hallobium: Analysis of the X-ray diffraction pattern. J Mol Biol. 1975;93:123–138. - PubMed
- Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. - PubMed
- Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: The helical hairpin hypothesis. Cell. 1981;23:411–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous