Arp2 links autophagic machinery with the actin cytoskeleton - PubMed (original) (raw)
Arp2 links autophagic machinery with the actin cytoskeleton
Iryna Monastyrska et al. Mol Biol Cell. 2008 May.
Abstract
Macroautophagy involves lysosomal/vacuolar elimination of long-lived proteins and entire organelles from the cytosol. The process begins with formation of a double-membrane vesicle that sequesters bulk cytoplasm, or a specific cargo destined for lysosomal/vacuolar delivery. The completed vesicle fuses with the lysosome/vacuole limiting membrane, releasing its content into the organelle lumen for subsequent degradation and recycling of the resulting macromolecules. A majority of the autophagy-related (Atg) proteins are required at the step of vesicle formation. The integral membrane protein Atg9 cycles between certain intracellular compartments and the vesicle nucleation site, presumably to supply membranes necessary for macroautophagic vesicle formation. In this study we have tracked the movement of Atg9 over time in living cells by using real-time fluorescence microscopy. Our results reveal that an actin-related protein, Arp2, briefly colocalizes with Atg9 and directly regulates the dynamics of Atg9 movement. We propose that proteins of the Arp2/3 complex regulate Atg9 transport for specific types of autophagy.
Figures
Figure 1.
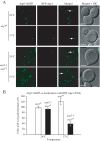
The arp2-1 mutant is defective in selective autophagy. (A) Precursor Ape1 processing is blocked in arp2-1 cells. The arp2-1 mutant cells were pulse-labeled for 10 min and subjected to a nonradioactive chase for 3 h at either 24°C (permissive temperature) or 37°C (nonpermissive temperature) in SMD medium, followed by an additional 3-h chase at 24°C as indicated. Rapamycin (Rap; 0.2 μg/ml) was added after pulse labeling, as indicated. Ape1 was immunoprecipitated and resolved by SDS-PAGE. The positions of precursor and mature Ape1 are marked. (B) Autophagy is delayed in the arp2-1 mutant. The wild-type, arp2-1, and _atg9_Δ strains carrying a plasmid expressing GFP-Atg8 [pGFPAUT7(414)] were grown in SMD at 24°C to early log phase, and then they were preincubated for 30 min either at 24 or 37°C and finally shifted to nitrogen-depleted (SD-N) medium to induce autophagy. Samples for the Western blot were taken before (0 h) and after 1.5 and 3 h of starvation. The numbers below each lane correspond to the percentage of cleaved (free) GFP. (C) Pexophagy is impaired in arp2-1 cells. The arp2-1 Pex14-GFP strain (IRA036) was grown at 24°C in oleate medium to induce massive peroxisome proliferation, then one portion of the culture was incubated for 3 h at 24°C and another portion at 37°C. Cells were then shifted to nitrogen starvation conditions to induce selective peroxisome degradation (pexophagy). Protein extracts were prepared at the time points indicated and resolved by SDS-PAGE, and after Western blot, membranes were probed with anti-GFP monoclonal antibody. Full-length chimeric Pex14-GFP (66 kDa) and free GFP (∼25 kDa) were detected as indicated. (D) The actin cytoskeleton morphology is altered in arp2-1 mutants. Fluorescence images of actin structure in wild-type and arp2-1 cells were collected after 3 h of incubation at 24 and 37°C. Cells were fixed, permeabilized, and subsequently incubated for 2 h at room temperature with TR phalloidin to stain the actin cytoskeleton.
Figure 2.
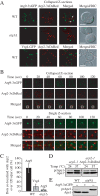
Anterograde transport of Atg9 is impaired in the arp2-1 mutant. (A) The _atg1_Δ and _arp2-1 atg1_Δ strains chromosomally-tagged with Atg9-3xGFP and RFP-Ape1, and carrying a centromeric plasmid (pATG1ts(414)) encoding a temperature-sensitive allele of ATG1 were grown to mid-log phase at 24 and 37°C and visualized by fluorescence microscopy. Arrows mark the site of the PAS. DIC, differential interference contrast. Scale bar, 2 μm. (B) Quantification of the Atg9-3xGFP and RFP-Ape1 colocalization from (A); colocalization was defined as one Atg9-3xGFP dot overlapping with the RFP-Ape1 signal. Approximately 50 cells expressing RFP-Ape1 were examined. The number of cells (40) where Atg9-3xGFP and RFP-Ape1 colocalized in the atg1 ts strain at 24°C was set to 100%. The values represent mean and standard deviation for 3 independent experiments.
Figure 3.
Movement of Atg9 is Arp2-dependent. Wild-type and arp2-1 cells were grown to mid-log phase and shifted to nonpermissive temperature for 30 min before visualization. Movement of Atg9-3xGFP patches was tracked by time-lapse fluorescence imaging as described in Materials and Methods. The presented images are still frames of the indicated boxed regions shown at 2× enlargement and taken with a time lapse of 2 s for 60-s duration. (A) In wild-type cells, the Atg9-3xGFP puncta rapidly changed their location accompanied by fusion or division. (B) In the arp2-1 conditional mutant, there was no displacement of the majority of Atg9-3xGFP puncta during 60 s. Bar, 2 μm. (C) The single Atg9-3xGFP puncta indicated in A and B were tracked in the wild-type Atg9-3xGFP puncta indicated in A and B were tracked in the wild-type versus arp2-1 strains. Spatial positions of the centers of puncta were determined in each frame (with 2-s intervals) of a movie using ImageJ software, and consecutive positions were connected by lines. Open, larger circles denote the first position of each punctum. Bar, 500 nm.
Figure 4.
Atg9 colocalizes with Arp2. (A) Atg9 and Arp2 were tracked in a single live cell in wild-type (IRA033) and _atg1_Δ (IRA037) cells expressing chromosomally tagged Atg9-3xGFP and TPI promoter-driven Arp2-3xDsRed, and Arp2 and chromosomally tagged Vrg4-GFP were monitored in wild-type cells (IRA039). Mid-log phase cells were analyzed by simultaneous two-color imaging combined in 4D (x,y,z,t). The RFP and GFP images were collected simultaneously with a time interval between Z-stacks acquisitions of 20 s for 120-s duration. A collapsed series of Z-sections is shown at the 60 s time point. Scale bar, 2 μm. (B) Still frames from a collapsed series of Z-sections and individual Z-sections at a single focal plane from a different cell demonstrate Atg9-3xGFP and Arp2-3xDsRed colocalization. The single Z-section images show the magnified view of the boxed area. (C) Quantification of the number of Atg9-3xGFP puncta colocalized with Arp2-3xDsRed in the wild-type and _atg1_Δ strains, and the number of Vrg4-GFP dots colocalized with Arp2-3xDsRed (n = 100). The number of Atg9 dots that colocalized with Arp2 in the wild-type strain per cell was set to 100%. (D) Arp2-3xDsRed is functional. arp2-1 cells expressing Arp2-DsRed or empty vector were grown to mid-log phase at 24°C, and then they were incubated for 3 h either at 24 or 37°C. Precursor Ape1 maturation was monitored by Western blot. The positions of precursor and mature Ape1 are indicated. (E) Atg9-3xGFP is functional. Wild-type cells or _atg9_Δ cells expressing Atg9-3xGFP or empty vector were grown to mid-log phase, and then they were examined by Western blot as described in C.
Figure 5.
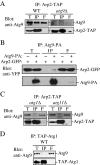
Atg9 binds to Arp2. (A) Atg9 is coimmunoprecipitated with Arp2. Either wild-type cells containing chromosomally TAP-tagged Arp2 (Arp2-TAP, WT) or _atg9_Δ cells with chromosomally tagged Arp2-TAP (IRA020) were used for affinity isolation as described in Materials and Methods. Eluted polypeptides were separated by SDS-PAGE and detected with anti-Atg9 antiserum. The same amounts of the total lysate (T), immunoprecipitate (IP), and flow through (F) were loaded per gel lane. (B) Arp2 is coimmunoprecipitated with Atg9. A strain expressing chromosomally tagged Atg9-protein A (FRY171) and plasmid-based Arp2-GFP or a control strain (SEY6210) only expressing plasmid-based Arp2-GFP and protein A driven by the CUP1 promoter were used for affinity isolation as described in A. Protein bands were detected with anti-YFP antibody. (C) Endogenous Atg9 is not coimmunoprecipitated with Arp2 in the absence of Atg1 or Atg11. The indicated _atg1_Δ (IRA023) or _atg11_Δ (IRA028) Arp2-tagged strains were used for affinity isolation as in A and probed with anti-Atg9 antiserum. (D) Atg9 is not coimmunoprecipitated with Atg1. A strain with chromosomally TAP-tagged Atg1 (UNY102) served as a control for Atg9 affinity isolation. Eluted polypeptides were separated by SDS-PAGE and visualized by immunoblotting as described in A.
Figure 6.
A mutant Atg9 that loses the ability to interact with Atg11 does not bind Arp2. (A) Endogenous Atg9 is coimmunoprecipitated with Arp2 in a wild-type strain but not in the absence of Atg11 or in a mutant of Atg9 that loses the ability to interact with Atg11. The indicated wild-type (ARP2-TAP, WT), _atg11_Δ (IRA028), or Atg9H192L (IRA020 harboring pATG9H192L) strains were used for affinity isolation as in Figure 5. (B) Mid-log phase cells were analyzed by simultaneous two-color imaging combined in 4D (x,y,z,t). The RFP and GFP images were collected simultaneously with a time interval between Z-stacks acquisitions of 10- for 120-s duration. Still frames from collapsed series Z-sections examine Atg9-3xGFP and Arp2-3xDsRed colocalization (indicated by arrows) in the wild-type Atg9-3xGFP or mutant Atg9H192L-3xGFP strain. Bar, 2 μm. (C) Quantification of the number of Atg9-3xGFP dots colocalized with Arp2-3xDsRed per cell (n = 79) in the wild-type versus Atg9H192L mutant strain. Fifteen of a total of 105 Atg9-3xGFP dots displayed in 13 frames taken during a 120-s time course colocalized with Arp2 in the wild-type strain; this number was set to 100%. *p < 0.05 value indicates that a statistically significantly higher number of Atg9-3xGFP dots colocalized with Arp2-3xDsRed compared with the mutant Atg9H192L-3xGFP.
Figure 7.
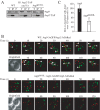
Atg9 interacts with arp2-1, which is defective in movement. (A) Atg9-3xGFP and arp2-1-3xDsRed colocalize. arp2-1 cells expressing chromosomally tagged Atg9-3xGFP and arp2-1-3xDsRed (JGY086) were grown to mid-log phase at 24°C, and then they were shifted to 37°C for 30 min. Samples were collected before and after the temperature shift, and then they were analyzed by microscopy as described in Materials and Methods. Colocalization of Atg9-3xGFP and arp2-1-3xDsRed in a single Z-section is indicated by arrows. (B) The arp2-1 protein interacts with Atg9. Cells (IRA038) expressing the chromosomally TAP-tagged arp2-1 protein were grown in SMD medium at 24°C, and then one aliquot of the culture was shifted to 37°C for 3 h. Cells were collected, and affinity isolation was performed as in Materials and Methods. The same amounts of total lysate (T), immunoprecipitate (IP), and flow through (F) were separated by SDS-PAGE and detected by anti-Atg9 antiserum. Bar, 2 μm. (C) Atg9 is not coimmunoprecipitated with protein A (PA) alone. Wild-type cells expressing CUP1 promoter-driven PA were used as a negative control for Atg9 interaction. Cells were cultured at 30°C and analyzed by affinity isolation as described in B. (D) arp2-1-DsRed is defective in movement at the nonpermissive temperature. Mid-log phase cells expressing arp2-1-3xDsRed (JGY086) were grown and imaged as in described in A with a time interval between Z-stack acquisitions of 10 s for 120-s duration. Still frames from collapsed series Z-sections examine arp2-1-3xDsRed movement at 24 and 37°C. Bar, 2 μm.
Figure 8.
The Arp2/3 complex is necessary for selective autophagy. (A) Precursor Ape1 maturation is blocked in the arp3-2 (RLY193), arc15-10 (Y4963), arc35-6 (BGY0809), and arc40-118 (BGY0883) strains. The cells were grown to mid-log phase at 24°C, and then they were incubated for 3 h either at 24 or 37°C. Precursor Ape1 maturation was monitored with pulse-chase radiolabeling followed by immunoprecipitation with antiserum to Ape1. The positions of precursor and mature Ape1 are indicated. (B) GFP-Atg8 processing is delayed in some strains with mutations in Arp2/3 complex subunits. The wild-type, arp3-2, arc15-10, arc35-6, and arc40-118 strains transformed with a GFP-Atg8 plasmid [pGFPAUT7(414) or pGFPAUT7(416)] were grown in SMD to mid-log phase at permissive temperature and preincubated for 30 min either at 24 or 37°C before being shifted to starvation conditions (SD-N). At the indicated time points, aliquots were taken and protein extracts were analyzed by Western blot by using anti-GFP antibody. The positions of GFP-Atg8 and free GFP are indicated along with the percentage of free GFP. (C) Mutations in Arp2/3 complex regulatory proteins block selective autophagy. The wild-type (SEY6210) and _atg9_Δ (JKY007) strains were grown to mid-log phase at 30°C, and protein extracts were prepared for controls. The pan1-3 (YAS1115), las17-11 (DDY1960), las17_Δ_WCA (DDY3045) and las17_Δ_WCA pan1_Δ_855-1480 (DDY2885) strains were grown to mid-log phase at 24°C, shifted to 37°C for 60 min, and examined by Western blot as described in A.
Similar articles
- Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast.
He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. He C, et al. J Cell Biol. 2006 Dec 18;175(6):925-35. doi: 10.1083/jcb.200606084. J Cell Biol. 2006. PMID: 17178909 Free PMC article. - Atg9 trafficking in autophagy-related pathways.
He C, Klionsky DJ. He C, et al. Autophagy. 2007 May-Jun;3(3):271-4. doi: 10.4161/auto.3912. Epub 2007 May 29. Autophagy. 2007. PMID: 17329962 - Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae.
Reggiori F, Klionsky DJ. Reggiori F, et al. J Cell Sci. 2006 Jul 15;119(Pt 14):2903-11. doi: 10.1242/jcs.03047. Epub 2006 Jun 20. J Cell Sci. 2006. PMID: 16787937 Free PMC article. - Atg9 trafficking in the yeast Saccharomyces cerevisiae.
Mari M, Reggiori F. Mari M, et al. Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review. - NPFs-mediated actin cytoskeleton: a new viewpoint on autophagy regulation.
Dong Y, Quan C. Dong Y, et al. Cell Commun Signal. 2024 Feb 12;22(1):111. doi: 10.1186/s12964-023-01444-2. Cell Commun Signal. 2024. PMID: 38347641 Free PMC article. Review.
Cited by
- Proximity-dependent biotinylation screening identifies NbHYPK as a novel interacting partner of ATG8 in plants.
Macharia MW, Tan WYZ, Das PP, Naqvi NI, Wong SM. Macharia MW, et al. BMC Plant Biol. 2019 Jul 19;19(1):326. doi: 10.1186/s12870-019-1930-8. BMC Plant Biol. 2019. PMID: 31324141 Free PMC article. - ARP2/3 complex associates with peroxisomes to participate in pexophagy in plants.
Martinek J, Cifrová P, Vosolsobě S, García-González J, Malínská K, Mauerová Z, Jelínková B, Krtková J, Sikorová L, Leaves I, Sparkes I, Schwarzerová K. Martinek J, et al. Nat Plants. 2023 Nov;9(11):1874-1889. doi: 10.1038/s41477-023-01542-6. Epub 2023 Oct 16. Nat Plants. 2023. PMID: 37845336 - Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy.
Wang K, Jin M, Liu X, Klionsky DJ. Wang K, et al. Autophagy. 2013 Nov 1;9(11):1828-36. doi: 10.4161/auto.26281. Epub 2013 Sep 6. Autophagy. 2013. PMID: 24025448 Free PMC article. - Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells.
Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, Rabinowich H. Han J, et al. J Biol Chem. 2008 Jul 11;283(28):19665-77. doi: 10.1074/jbc.M710169200. Epub 2008 Mar 28. J Biol Chem. 2008. PMID: 18375389 Free PMC article. - Actin network evolution as a key driver of eukaryotic diversification.
Velle KB, Swafford AJM, Garner E, Fritz-Laylin LK. Velle KB, et al. J Cell Sci. 2024 Aug 1;137(15):jcs261660. doi: 10.1242/jcs.261660. Epub 2024 Aug 9. J Cell Sci. 2024. PMID: 39120594 Free PMC article. Review.
References
- Aplin A., Jasionowski T., Tuttle D. L., Lenk S. E., Dunn W. A., Jr Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J. Cell. Physiol. 1992;152:458–466. - PubMed
- Ayscough K. R. Coupling actin dynamics to the endocytic process in Saccharomyces cerevisiae. Protoplasma. 2005;226:81–88. - PubMed
- Birmingham C. L., Brumell J. H. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials