SPIKE--a database, visualization and analysis tool of cellular signaling pathways - PubMed (original) (raw)
Comparative Study
doi: 10.1186/1471-2105-9-110.
Rita Vesterman, Nira Amit, Igor Ulitsky, Idan Zohar, Mali Weisz, Gilad Mass, Nir Orlev, Giora Sternberg, Ran Blekhman, Jackie Assa, Yosef Shiloh, Ron Shamir
Affiliations
- PMID: 18289391
- PMCID: PMC2263022
- DOI: 10.1186/1471-2105-9-110
Comparative Study
SPIKE--a database, visualization and analysis tool of cellular signaling pathways
Ran Elkon et al. BMC Bioinformatics. 2008.
Abstract
Background: Biological signaling pathways that govern cellular physiology form an intricate web of tightly regulated interlocking processes. Data on these regulatory networks are accumulating at an unprecedented pace. The assimilation, visualization and interpretation of these data have become a major challenge in biological research, and once met, will greatly boost our ability to understand cell functioning on a systems level.
Results: To cope with this challenge, we are developing the SPIKE knowledge-base of signaling pathways. SPIKE contains three main software components: 1) A database (DB) of biological signaling pathways. Carefully curated information from the literature and data from large public sources constitute distinct tiers of the DB. 2) A visualization package that allows interactive graphic representations of regulatory interactions stored in the DB and superposition of functional genomic and proteomic data on the maps. 3) An algorithmic inference engine that analyzes the networks for novel functional interplays between network components.SPIKE is designed and implemented as a community tool and therefore provides a user-friendly interface that allows registered users to upload data to SPIKE DB. Our vision is that the DB will be populated by a distributed and highly collaborative effort undertaken by multiple groups in the research community, where each group contributes data in its field of expertise.
Conclusion: The integrated capabilities of SPIKE make it a powerful platform for the analysis of signaling networks and the integration of knowledge on such networks with omics data.
Figures
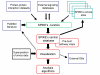
Figure 1
An overview of SPIKE. This scheme describes the key components of the system and information flow. SPIKE's three main software components are: 1) A DB of signaling pathways, containing, in distinct tiers, carefully ascertained information uploaded by SPIKE users and supervised by the curators, data from external signaling DBs (e.g., KEGG, Reactome, IntAct), and wide-scale human protein-protein interaction datasets [21–24]. In SPIKE's decentralized architecture, a copy of the DB is installed along with the software in each participating research lab, and these remote site DBs are periodically synchronized with the central DB. The information stored in the SPIKE DB can be readily used by other KBs. 2) A visualization package that allows interactive representation of selected regulatory interactions from the DB, dynamic layout and navigation through the displayed networks, and superposition of high-throughput genomic and proteomic data. Information displayed in the maps is linked to external DBs (e.g., Entrez Genes, PubMed). Prearranged maps for key signaling networks (e.g., p53, apoptosis, cell cycle, ATM, MAPK) were built and are posted at SPIKE website. 3) An algorithmic engine that performs various network analyses, aimed at discovering novel functional interplays between network components.
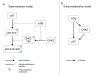
Figure 2
SPIKE modeling scheme. State transition scheme vs. entity-relationship scheme. (A). The 'state transition' scheme regards different post-translational modified versions of a protein as separate entities (or as different 'states' of a protein), visually represents them as distinct nodes in the map, and aims at tracing the transition between these states. (B). In contrast, the 'entity-relationship' scheme views all these entities as one protein, and focuses on the regulatory effects between different proteins within a signaling network. In a state transition scheme, representation of signaling pathways could become extremely complex due to combinatorial growth of possible states. An entity-relationship scheme is much simpler, having all these states represented by only one entity, but at the price of not taking into account temporal order constraints on the state transition reactions in cases where such constraints exist.
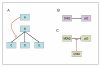
Figure 3
Regulation as target of another regulation. (A). The ability to define a regulation as a target of another regulation is helpful in cases where the effect of regulator A on target B is transmitted to some but not all targets of B. In this schematic example, A specifically inhibits the B-mediated activation of C. (B). A schematic representation of p53 activation by CHK2 [31]. (C). A more specific representation: here, the information that the activation of p53 is achieved by CHK2 interfering with the inhibition of p53 by MDM2 is explicitly represented. This interference is achieved by CHK2 phosphorylation of p53 [31], and therefore the 'physical target' attribute of this regulation is p53.
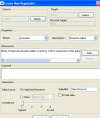
Figure 4
Adding new information to SPIKE DB. Submission forms allow privileged users to upload data to the DB. Here, the form for submission of a new regulation is shown. The user specifies the source, target and effect of the regulation. Additional regulation attributes include the biochemical mechanism, one or more supporting references, the submitter, and the quality level ascribed to the data. Submitted data can be flagged as private, in which case the data will be visible only to the user who uploaded it.
Figure 5
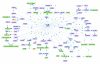
SPIKE visualization of signaling networks. The different types of biological entities are represented in SPIKE maps by nodes of different colors: violet nodes correspond to protein-coding genes, turquoise nodes to non protein-coding genes (e.g., micro-RNAs, rRNAs, tRNAs), green nodes to protein complexes, and yellow nodes to protein families. Small molecules are displayed in orange (not included in this map). Blue directed edges represent regulations: arrows correspond to activation, T shape edges to inhibition, and open circles denote regulations whose effect is still not clear (e.g., ATM was reported to phosphorylate MCM2, but the effect of this modification is not known yet). Blue undirected edges represent protein-protein interactions (not included in this map). Green edges represent containment relations between nodes (e.g., a complex and its components). Red and green dots within a node indicate that not all regulation and containment relations stored in the DB for the node are displayed in the map. This map represents the ATM-regulated network which is set off by the cell in response to double-strand breaks in the DNA.
Figure 6
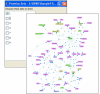
Superposition of 'omics' data on maps. By allowing the user to superimpose functional genomic or proteomic data on pathway maps, SPIKE facilitates improved interpretation of such datasets. The color of the bar above each gene's node indicates its response. Upregulated genes are shades of red: the darker the red the greater the fold of induction. Similarly, downregulated genes are in shades of green. Genes whose expression was not changed are yellow, and genes for which data are not available (e.g., genes not present on the microarray) are grey. In this example, superimposing gene expression data measured in human lymphocyte cells exposed to ionizing radiation on the signaling map shows the activation of the p53 network (top-left part of the network) and the down-regulation of genes that function in the cell cycle G2-M phase transition (bottom-right part of the network). Superimposing the data on this map clearly shows that a large fraction of the p53 network was activated in the analyzed condition. SPIKE also allows superimposition of clustering data: any partition of the genes into groups (e.g., according to GO annotation, cellular compartment, clustering, or user-specific definitions) can be viewed on the pathway map. The color scheme can be adjusted by the user.
Figure 7
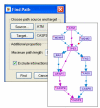
Path finding. The user specifies the source, the target and the maximal number of edges in a path connecting them. By default, un-directed edges (that is, protein-protein interactions) are excluded from the analysis. In this example, pathways linking ATM to CASP3, a major effector of apoptosis, are sought. All paths that meet the length constraint are displayed with the shortest paths highlighted.
Figure 8
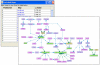
Interconnections within a set of genes/proteins. Interconnections among nodes within a specified target set of genes or proteins (e.g., sets obtained by cluster analysis applied to gene expression microarray data) are displayed. This figure shows the interconnections within a cluster of genes that were induced by LPS treatment, including single intermediate nodes not contained in the original cluster. Nodes corresponding to genes contained in the input cluster are highlighted in pink.
Figure 9
Enriched maps. Signaling maps enriched for nodes included in target sets of genes/proteins are sought. Maps that are significantly enriched (beyond a specified p-value threshold) are listed on the results table. Clicking on a map's link in this table opens the enriched map, in which nodes contained in the original input set are highlighted.
Similar articles
- Biological Network Inference and analysis using SEBINI and CABIN.
Taylor R, Singhal M. Taylor R, et al. Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review. - BEL Commons: an environment for exploration and analysis of networks encoded in Biological Expression Language.
Hoyt CT, Domingo-Fernández D, Hofmann-Apitius M. Hoyt CT, et al. Database (Oxford). 2018 Jan 1;2018:bay126. doi: 10.1093/database/bay126. Database (Oxford). 2018. PMID: 30576488 Free PMC article. - ProteoLens: a visual analytic tool for multi-scale database-driven biological network data mining.
Huan T, Sivachenko AY, Harrison SH, Chen JY. Huan T, et al. BMC Bioinformatics. 2008 Aug 12;9 Suppl 9(Suppl 9):S5. doi: 10.1186/1471-2105-9-S9-S5. BMC Bioinformatics. 2008. PMID: 18793469 Free PMC article. - SPIKE: a database of highly curated human signaling pathways.
Paz A, Brownstein Z, Ber Y, Bialik S, David E, Sagir D, Ulitsky I, Elkon R, Kimchi A, Avraham KB, Shiloh Y, Shamir R. Paz A, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D793-9. doi: 10.1093/nar/gkq1167. Epub 2010 Nov 19. Nucleic Acids Res. 2011. PMID: 21097778 Free PMC article. - Network portal: a database for storage, analysis and visualization of biological networks.
Turkarslan S, Wurtmann EJ, Wu WJ, Jiang N, Bare JC, Foley K, Reiss DJ, Novichkov P, Baliga NS. Turkarslan S, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D184-90. doi: 10.1093/nar/gkt1190. Epub 2013 Nov 23. Nucleic Acids Res. 2014. PMID: 24271392 Free PMC article.
Cited by
- Integrated proteomic, transcriptomic, and biological network analysis of breast carcinoma reveals molecular features of tumorigenesis and clinical relapse.
Imielinski M, Cha S, Rejtar T, Richardson EA, Karger BL, Sgroi DC. Imielinski M, et al. Mol Cell Proteomics. 2012 Jun;11(6):M111.014910. doi: 10.1074/mcp.M111.014910. Epub 2012 Jan 12. Mol Cell Proteomics. 2012. PMID: 22240506 Free PMC article. - Coordination between cell cycle progression and cell fate decision by the p53 and E2F1 pathways in response to DNA damage.
Zhang XP, Liu F, Wang W. Zhang XP, et al. J Biol Chem. 2010 Oct 8;285(41):31571-80. doi: 10.1074/jbc.M110.134650. Epub 2010 Aug 4. J Biol Chem. 2010. PMID: 20685653 Free PMC article. - The OncoFinder algorithm for minimizing the errors introduced by the high-throughput methods of transcriptome analysis.
Buzdin AA, Zhavoronkov AA, Korzinkin MB, Roumiantsev SA, Aliper AM, Venkova LS, Smirnov PY, Borisov NM. Buzdin AA, et al. Front Mol Biosci. 2014 Aug 26;1:8. doi: 10.3389/fmolb.2014.00008. eCollection 2014. Front Mol Biosci. 2014. PMID: 25988149 Free PMC article. - Hypergraph-based connectivity measures for signaling pathway topologies.
Franzese N, Groce A, Murali TM, Ritz A. Franzese N, et al. PLoS Comput Biol. 2019 Oct 25;15(10):e1007384. doi: 10.1371/journal.pcbi.1007384. eCollection 2019 Oct. PLoS Comput Biol. 2019. PMID: 31652258 Free PMC article. - Expander: from expression microarrays to networks and functions.
Ulitsky I, Maron-Katz A, Shavit S, Sagir D, Linhart C, Elkon R, Tanay A, Sharan R, Shiloh Y, Shamir R. Ulitsky I, et al. Nat Protoc. 2010 Feb;5(2):303-22. doi: 10.1038/nprot.2009.230. Epub 2010 Jan 28. Nat Protoc. 2010. PMID: 20134430
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous