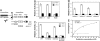Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways - PubMed (original) (raw)
. 2008 Mar 4;105(9):3368-73.
doi: 10.1073/pnas.0711591105. Epub 2008 Feb 25.
Sarah Linke, José M Dias, Xiaowei Zheng, Katarina Gradin, Tristan P Wallis, Brett R Hamilton, Maria Gustafsson, Jorge L Ruas, Sarah Wilkins, Rebecca L Bilton, Kerstin Brismar, Murray L Whitelaw, Teresa Pereira, Jeffrey J Gorman, Johan Ericson, Daniel J Peet, Urban Lendahl, Lorenz Poellinger
Affiliations
- PMID: 18299578
- PMCID: PMC2265116
- DOI: 10.1073/pnas.0711591105
Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways
Xiaofeng Zheng et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Cells adapt to hypoxia by a cellular response, where hypoxia-inducible factor 1alpha (HIF-1alpha) becomes stabilized and directly activates transcription of downstream genes. In addition to this "canonical" response, certain aspects of the pathway require integration with Notch signaling, i.e., HIF-1alpha can interact with the Notch intracellular domain (ICD) to augment the Notch downstream response. In this work, we demonstrate an additional level of complexity in this cross-talk: factor-inhibiting HIF-1 (FIH-1) regulates not only HIF activity, but also the Notch signaling output and, in addition, plays a role in how Notch signaling modulates the hypoxic response. We show that FIH-1 hydroxylates Notch ICD at two residues (N(1945) and N(2012)) that are critical for the function of Notch ICD as a transactivator within cells and during neurogenesis and myogenesis in vivo. FIH-1 negatively regulates Notch activity and accelerates myogenic differentiation. In its modulation of the hypoxic response, Notch ICD enhances recruitment of HIF-1alpha to its target promoters and derepresses HIF-1alpha function. Addition of FIH-1, which has a higher affinity for Notch ICD than for HIF-1alpha, abrogates the derepression, suggesting that Notch ICD sequesters FIH-1 away from HIF-1alpha. In conclusion, the data reveal posttranslational modification of the activated form of the Notch receptor and an intricate mode of cross-coupling between the Notch and hypoxia signaling pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Hydroxylation of Notch ICD at 2 asparagine residues by FIH-1. (A) Schematic diagram showing key features of Notch. (B) Notch 1 ICD, ANK 1–7, and RAM proteins were analyzed in a 14CO2 capture assay. Data are mean relative activity ± range (n = 2) and are representative of >3 independent experiments. (C) Purified wild-type (WT) or Asn mutant ANK 1–7 proteins or (D) mNotch 1–4 ANK 1–7 proteins were compared for their abilities to promote FIH-1-mediated 2-oxoglutarate turnover. Data are presented as mean ± range (n = 2) and are representative of three independent experiments. (E) Kinetic CO2 capture assay with varying HIF-CAD or mNotch1 substrate concentrations, with data fitted to a hyperbolic curve using PRISM software. The HIF-CAD apparent _K_m is 36 μM, and the apparent _V_max is 4.7 nmol/mg per min, whereas Notch hydroxylation rate is near-maximal (≈0.7 nmol/mg per min), and the apparent _K_m cannot be determined accurately (<4 μM). Data are mean nmol/mg per min ± range (_n_ = 2), representative of >3 independent experiments.
Fig. 2.
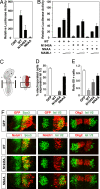
Mutation of Notch 1 ICD N1945 and N2012 reduces the transactivation potency and impairs Notch-mediated repression of neuronal differentiation. (A) Notch N1945/N2012 is a very weak transcriptional activator. P19 cells were transfected with 0.5 μg of a CSL-driven luciferase plasmid and 100 ng of CMX or plasmids encoding wild-type (WT), N1945A, or N1945A/N2012A (NNAA) mutant ICD. (B) Activity of Notch Asn mutants is rescued by expression of MAML1. HeLa cells were transfected with 0.5 μg of 12×CSL-Luc plasmid together with 100 ng of plasmids encoding WT or mutant forms of Notch 1 ICD in the presence of increasing concentrations of an expression vector (50 ng and 200 ng) encoding MAML1. Data are presented as luciferase activity relative to cells transfected with 12×CSL-Luc. Values represent the mean ± SE of three independent experiments performed in duplicate. (C) Schematic representation of a harvested chicken embryo after electroporation. Transverse sections at the forelimb level (dashed line) were analyzed. In the electroporated spinal cord, only one of the sides was electroporated (red region). The analysis was focused on the most ventral region of the spinal cord (boxed region). Quantification of the effect on motor neuron generation after expression of GFP or WT; N1945A, or NNAA Notch1 ICD is shown. The percentage of cells electroporated with the indicated constructs and expressing the postmitotic motor neuron marker Isl1/2 (D), and the ratio of the total number of motor neurons between the electroporated vs. control (el/co) side (E) were determined. (F) Effect of wild-type and mutant forms of Notch on neurogenesis in the ventral spinal cord. Embryos were electroporated with GFP or with wild-type or mutant Notch1 ICD as indicated. The ability of the different constructs to maintain cells in a progenitor stage or to repress motor neuron generation was analyzed by immunohistochemistry using antibodies against Sox3 or Isl1/2, respectively. Expression of the different constructs did not affect the motor neuron progenitor pool expressing Olig2 (F), arguing that the observed role of Notch does not result from progenitor pool depletion. +, electroporated side; −, control side.
Fig. 3.
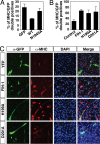
FIH-1 increases myogenic differentiation. (A) Quantification of the percentage of GFP-positive cells (expressing either GFP, Notch 1 ICD-GFP, or Notch 1 ICD N1945A-GFP) that are also MHC-positive. (B) Quantification of the percentage of YFP-positive cells that are also MHC-positive (see below). (C) Differentiated (MHC-positive, red) C2C12 cells costained for expression of YFP, FIH-1-YFP, FIH-1H199A-YFP, or FIH-1D201A-YFP (green).
Fig. 4.
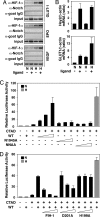
Notch ICD induces nuclear accumulation of FIH-1. (A) 293A cells were transfected with 0.4 μg of YFP-fused FIH-1 alone or together with 0.6 μg of plasmids encoding wild-type Notch ICD (WT) or ICD N1945A/N2012A (NNAA). Cells were analyzed by confocal microscopy, and intracellular distribution was categorized as described in ref. . Nuclear compartment was identified by DAPI staining. (B) Notch affects cellular localization of FIH-1 in vivo. Chick neural tubes were electroporated with FIH-1 alone or together with wild-type or NNAA mutant Notch1 ICD. Cellular localization of FIH-1 was determined by immunohistochemistry (α-FIH-1). The zoom panels on each column correspond to magnification of the dashed yellow boxes. (C) Levels of FIH-1 mRNA in cells transfected with control siRNA and FIH-1-specific siRNA and analyzed by quantitative RT-PCR. (D) siRNA targeting FIH-1 produces a modest effect on Notch-dependent luciferase activity. 293T cells stably expressing full-length Notch and plated on IgG (ligand −) or Jagged-1-coated plates (ligand +) were first transfected with 100 nM negative control or a FIH-1-specific siRNA and 24 h later were transfected again with 0.5 μg of 12×CSL-Luc plasmid. (E) Inhibition of FIH-1 expression by siRNA mediates derepression of HIF-1α CAD function. Twenty-four hours after transfection with siRNAs, a GAL4-driven luciferase plasmid (0.5 μg) and an expression plasmid encoding GAL4-fused HIF-1α C-TAD (50 ng) were transfected. Data are presented as luciferase activity relative to cells transfected with reporter gene alone. Values represent the mean ± SE of three independent experiments performed in duplicate.
Fig. 5.
Activation of Notch increases binding of HIF-1α to target genes and causes derepression of HIF-1α function. (A) Binding of HIF-1α to target gene HREs by ChIP analysis in the absence or presence of activated Notch. C2C12 cells stably expressing full-length Notch were cultured in the presence of control 293T cells (ligand −) or 293T cells stably expressing Notch ligand Serrate-1 (ligand +) and kept at normoxia (N) or exposed to hypoxia (H). ChIP assays were performed using the indicated antibodies, and RT-PCR analysis of target gene HREs was carried out. (B) Activation of the Notch pathway increases HIF-1 target gene expression. 293T cells stably expressing full-length Notch were cultured on plates coated with IgG (ligand −) or Jagged-1 (ligand +) and kept at normoxia (N) or hypoxia (H). Total RNA was prepared, and quantitative RT-PCR was performed. (C) Notch ICD derepresses HIF-1α CAD function. P19 cells were transfected with 0.5 μg of a GAL4-driven luciferase plasmid and 50 ng of GAL-fused HIF-1α C-TAD expression plasmid in the presence of increasing amounts (50, 100, and 200 ng) of plasmids encoding wild-type or Asn mutant Notch1 ICD. (D) Expression of wild-type FIH-1 restores repression of CAD function at normoxia. P19 cells were transfected with 0.5 μg of a GAL4-driven luciferase plasmid together with a GAL-fused HIF-1α CAD expression plasmid (50 ng) and a plasmid encoding wild-type Notch 1 ICD (200 ng) in the presence of increasing amounts (50, 100, 200 ng) of plasmids encoding wild-type or mutant (D201A and D199A) FIH-1. Data are presented as luciferase activity relative to cells transfected with GAL4-Luc. Values represent the mean ± S.E. of three independent experiments performed in duplicate (C and D).
Similar articles
- Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor.
Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, Cook KM, Cockman ME, Lancaster DE, Kessler BM, Oldham NJ, Ratcliffe PJ, Schofield CJ. Coleman ML, et al. J Biol Chem. 2007 Aug 17;282(33):24027-38. doi: 10.1074/jbc.M704102200. Epub 2007 Jun 15. J Biol Chem. 2007. PMID: 17573339 - Factor inhibiting HIF (FIH) recognizes distinct molecular features within hypoxia-inducible factor-α (HIF-α) versus ankyrin repeat substrates.
Wilkins SE, Karttunen S, Hampton-Smith RJ, Murchland I, Chapman-Smith A, Peet DJ. Wilkins SE, et al. J Biol Chem. 2012 Mar 16;287(12):8769-81. doi: 10.1074/jbc.M111.294678. Epub 2012 Jan 23. J Biol Chem. 2012. PMID: 22270367 Free PMC article. - Differences in hydroxylation and binding of Notch and HIF-1alpha demonstrate substrate selectivity for factor inhibiting HIF-1 (FIH-1).
Wilkins SE, Hyvärinen J, Chicher J, Gorman JJ, Peet DJ, Bilton RL, Koivunen P. Wilkins SE, et al. Int J Biochem Cell Biol. 2009 Jul;41(7):1563-71. doi: 10.1016/j.biocel.2009.01.005. Epub 2009 Jan 20. Int J Biochem Cell Biol. 2009. PMID: 19401150 - Regulation of Transactivation at C-TAD Domain of HIF-1_α_ by Factor-Inhibiting HIF-1_α_ (FIH-1): A Potential Target for Therapeutic Intervention in Cancer.
Rani S, Roy S, Singh M, Kaithwas G. Rani S, et al. Oxid Med Cell Longev. 2022 May 10;2022:2407223. doi: 10.1155/2022/2407223. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35592530 Free PMC article. Review. - The interplay between the cellular hypoxic response and Notch signaling.
Landor SK, Lendahl U. Landor SK, et al. Exp Cell Res. 2017 Jul 15;356(2):146-151. doi: 10.1016/j.yexcr.2017.04.030. Epub 2017 Apr 26. Exp Cell Res. 2017. PMID: 28456549 Review.
Cited by
- Kuwanon V inhibits proliferation, promotes cell survival and increases neurogenesis of neural stem cells.
Kong SY, Park MH, Lee M, Kim JO, Lee HR, Han BW, Svendsen CN, Sung SH, Kim HJ. Kong SY, et al. PLoS One. 2015 Feb 23;10(2):e0118188. doi: 10.1371/journal.pone.0118188. eCollection 2015. PLoS One. 2015. PMID: 25706719 Free PMC article. - Donor treatment with a PHD-inhibitor activating HIFs prevents graft injury and prolongs survival in an allogenic kidney transplant model.
Bernhardt WM, Gottmann U, Doyon F, Buchholz B, Campean V, Schödel J, Reisenbuechler A, Klaus S, Arend M, Flippin L, Willam C, Wiesener MS, Yard B, Warnecke C, Eckardt KU. Bernhardt WM, et al. Proc Natl Acad Sci U S A. 2009 Dec 15;106(50):21276-81. doi: 10.1073/pnas.0903978106. Epub 2009 Nov 23. Proc Natl Acad Sci U S A. 2009. PMID: 19934037 Free PMC article. - Action Sites and Clinical Application of HIF-1α Inhibitors.
Xu R, Wang F, Yang H, Wang Z. Xu R, et al. Molecules. 2022 May 26;27(11):3426. doi: 10.3390/molecules27113426. Molecules. 2022. PMID: 35684364 Free PMC article. Review. - Not(ch) just development: Notch signalling in the adult brain.
Ables JL, Breunig JJ, Eisch AJ, Rakic P. Ables JL, et al. Nat Rev Neurosci. 2011 May;12(5):269-83. doi: 10.1038/nrn3024. Nat Rev Neurosci. 2011. PMID: 21505516 Free PMC article. Review. - Metabolites as drivers and targets in rheumatoid arthritis.
Hanlon MM, Canavan M, Barker BE, Fearon U. Hanlon MM, et al. Clin Exp Immunol. 2022 Jun 11;208(2):167-180. doi: 10.1093/cei/uxab021. Clin Exp Immunol. 2022. PMID: 35020864 Free PMC article.
References
- Semenza GL. Life with oxygen. Science. 2007;318:62–64. - PubMed
- Hewitson KS, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2002;277:26351–26355. - PubMed
- Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: A hypoxic switch. Science. 2002;295:858–861. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases