Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids - PubMed (original) (raw)
Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids
Sylvain L'Espérance et al. BMC Genomics. 2008.
Abstract
Background: Chemotherapy (CT) resistance in ovarian cancer (OC) is broad and encompasses diverse unrelated drugs, suggesting more than one mechanism of resistance. To better understand the molecular mechanisms controlling the immediate response of OC cells to CT exposure, we have performed gene expression profiling in spheroid cultures derived from six OC cell lines (OVCAR3, SKOV3, TOV-112, TOV-21, OV-90 and TOV-155), following treatment with 10,0 microM cisplatin, 2,5 microM paclitaxel or 5,0 microM topotecan for 72 hours.
Results: Exposure of OC spheroids to these CT drugs resulted in differential expression of genes associated with cell growth and proliferation, cellular assembly and organization, cell death, cell cycle control and cell signaling. Genes, functionally involved in DNA repair, DNA replication and cell cycle arrest were mostly overexpressed, while genes implicated in metabolism (especially lipid metabolism), signal transduction, immune and inflammatory response, transport, transcription regulation and protein biosynthesis, were commonly suppressed following all treatments. Cisplatin and topotecan treatments triggered similar alterations in gene and pathway expression patterns, while paclitaxel action was mainly associated with induction of genes and pathways linked to cellular assembly and organization (including numerous tubulin genes), cell death and protein synthesis. The microarray data were further confirmed by pathway and network analyses.
Conclusion: Most alterations in gene expression were directly related to mechanisms of the cytotoxics actions in OC spheroids. However, the induction of genes linked to mechanisms of DNA replication and repair in cisplatin- and topotecan-treated OC spheroids could be associated with immediate adaptive response to treatment. Similarly, overexpression of different tubulin genes upon exposure to paclitaxel could represent an early compensatory effect to this drug action. Finally, multicellular growth conditions that are known to alter gene expression (including cell adhesion and cytoskeleton organization), could substantially contribute in reducing the initial effectiveness of CT drugs in OC spheroids. Results described in this study underscore the potential of the microarray technology for unraveling the complex mechanisms of CT drugs actions in OC spheroids and early cellular response to treatment.
Figures
Figure 1
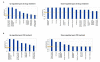
Functional analysis for a dataset of differentially expressed genes (≥1.5 fold) in OC spheroids following CT drugs treatments. A. Functional analysis following all drugs (cisplatin, topotecan and paclitaxel) treatment, B. Functional analysis following cisplatin treatment. Top functions that meet a _p_-value cutoff of 0.05 are displayed.
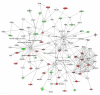
Figure 2
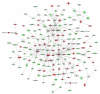
Network analysis of dynamic gene expression in OC spheroids based on the 1.5-fold common gene expression list obtained following treatment with all CT drugs used (cisplatin, topotecan and paclitaxel). The five top-scoring networks were merged and are displayed graphically as node (genes/gene product) and edges (the biological relationships between the nodes). Intensity of the node color indicates the degree of up- (red) or downregulation (green). Nodes are displayed using various shapes that represent the functional class of the gene product (square, cytokine, vertical oval, transmembrane receptor, rectangle, nuclear receptor, diamond, enzyme, rhomboid, transporter, hexagon, translation factor, horizontal oval, transcription factor, circle, other). Edges are displayed with various labels that describe the nature of relationship between the nodes: ---- binding only, → acts on. The length of an edge reflect the evidence supporting that node-to-node relationship, in that edges supported by article from literature are shorter. Dotted edges represent indirect interaction.
Figure 3
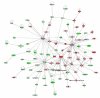
Network analysis of dynamic gene expression in OC spheroids based on the 1.5-fold common gene expression list obtained following cisplatin treatment. The three top-scoring networks were merged and are displayed graphically as nodes (genes/gene products) and edges (the biological relationships between the nodes). Figure legends are as described in Fig. 2.
Figure 4
Functional analysis for a dataset of differentially expressed genes (≥1.5 fold) in OC spheroids following CT drugs treatments. A. Functional analysis following topotecan treatment, B. Functional analysis following paclitaxel treatment. Top functions that meet a _p_-value cutoff of 0.05 are displayed.
Figure 5
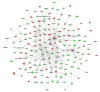
Network analysis of dynamic gene expression in OC spheroids based on the 1.5-fold common gene expression list obtained following topotecan treatment. The five top-scoring networks were merged and are displayed graphically as nodes (genes/gene products) and edges (the biological relationships between the nodes). Figure legends are as described in Fig. 2.
Figure 6
Network analysis of dynamic gene expression in OC spheroids based on the 1.5-fold common gene expression list obtained following paclitaxel treatment. The three top-scoring networks were merged and are displayed graphically as nodes (genes/gene products) and edges (the biological relationships between the nodes). Figure legends are as described in Fig. 2.
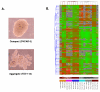
Figure 7
A. Example images of compact and aggregate spheroid structures derived from OC cells. B. Hierarchical clustering of OC spheroids following treatment with all used drugs (cisplatin, topotecan and paclitaxel (taxol)), that discriminates between compact spheroids and aggregates. A subset of candidate genes were initially obtained by filtering on signal intensity (2-fold), retaining 527 genes. One-way ANOVA parametric test (Welch _t_-test, variances not assumed equal, p ≤ 0.03) further selected 85 genes. Clustering analysis based on the 85 gene list was performed using the standard Condition Tree algorithm provided in GeneSpring. The mean appears grey, whereas red signifies up-regulation, and green signifies down-regulation (see legend bar). Compact spheroids are indicated in brown, aggregates are indicated in grey. Each cell line is indicated with different color.
Similar articles
- HE4 promotes collateral resistance to cisplatin and paclitaxel in ovarian cancer cells.
Ribeiro JR, Schorl C, Yano N, Romano N, Kim KK, Singh RK, Moore RG. Ribeiro JR, et al. J Ovarian Res. 2016 May 17;9(1):28. doi: 10.1186/s13048-016-0240-0. J Ovarian Res. 2016. PMID: 27184254 Free PMC article. - Long non-coding RNA Linc00312 modulates the sensitivity of ovarian cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway.
Zhang C, Wang M, Shi C, Shi F, Pei C. Zhang C, et al. Biosci Trends. 2018 Jul 17;12(3):309-316. doi: 10.5582/bst.2018.01052. Epub 2018 Jun 28. Biosci Trends. 2018. PMID: 29952351 - Bioinformatics analysis of mRNA and miRNA microarray to identify the key miRNA-mRNA pairs in cisplatin-resistant ovarian cancer.
Xue B, Li S, Jin X, Liu L. Xue B, et al. BMC Cancer. 2021 Apr 23;21(1):452. doi: 10.1186/s12885-021-08166-z. BMC Cancer. 2021. PMID: 33892654 Free PMC article. - A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.
Forbes C, Shirran L, Bagnall AM, Duffy S, ter Riet G. Forbes C, et al. Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100 Review. - Chemotherapy resistance in ovarian cancer: new molecular perspectives.
Coukos G, Rubin SC. Coukos G, et al. Obstet Gynecol. 1998 May;91(5 Pt 1):783-92. doi: 10.1016/s0029-7844(98)00054-4. Obstet Gynecol. 1998. PMID: 9572231 Review.
Cited by
- Role of AMIGO2 in cancer progression: Novel insights (Review).
Tian Z, Zhou D, Jiang R, Zhou B. Tian Z, et al. Oncol Lett. 2024 Jul 12;28(3):434. doi: 10.3892/ol.2024.14567. eCollection 2024 Sep. Oncol Lett. 2024. PMID: 39049987 Free PMC article. Review. - Norepinephrine induces anoikis resistance in high-grade serous ovarian cancer precursor cells.
Reavis HD, Gysler SM, McKenney GB, Knarr M, Lusk HJ, Rawat P, Rendulich HS, Mitchell MA, Berger DS, Moon JS, Ryu S, Mainigi M, Iwanicki MP, Hoon DS, Sanchez LM, Drapkin R. Reavis HD, et al. JCI Insight. 2024 Mar 8;9(5):e170961. doi: 10.1172/jci.insight.170961. JCI Insight. 2024. PMID: 38271085 Free PMC article. - Transcriptional Landscape of 3D vs. 2D Ovarian Cancer Cell Models.
Kerslake R, Belay B, Panfilov S, Hall M, Kyrou I, Randeva HS, Hyttinen J, Karteris E, Sisu C. Kerslake R, et al. Cancers (Basel). 2023 Jun 26;15(13):3350. doi: 10.3390/cancers15133350. Cancers (Basel). 2023. PMID: 37444459 Free PMC article. - Synergistic Effect of Anethole and Platinum Drug Cisplatin against Oral Cancer Cell Growth and Migration by Inhibiting MAPKase, Beta-Catenin, and NF-κB Pathways.
Semlali A, Ajala I, Beji S, Al-Zharani MM, Rouabhia M. Semlali A, et al. Pharmaceuticals (Basel). 2023 May 5;16(5):700. doi: 10.3390/ph16050700. Pharmaceuticals (Basel). 2023. PMID: 37242484 Free PMC article. - MicroRNA-dependent inhibition of WEE1 controls cancer stem-like characteristics and malignant behavior in ovarian cancer.
Cho JG, Kim SW, Lee A, Jeong HN, Yun E, Choi J, Jeong SJ, Chang W, Oh S, Yoo KH, Lee JB, Yoon S, Lee MS, Park JH, Jung MH, Kim SW, Kim KH, Suh DS, Choi KU, Choi J, Kim J, Kwon BS. Cho JG, et al. Mol Ther Nucleic Acids. 2022 Aug 24;29:803-822. doi: 10.1016/j.omtn.2022.08.028. eCollection 2022 Sep 13. Mol Ther Nucleic Acids. 2022. PMID: 36159587 Free PMC article.
References
- Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics 2001. CA Cancer J Clin. 2001;51:15–36. - PubMed
- du Bois A, Quinn M, Thigpen T, Vermorken J, Avall-Lundqvist E, Bookman M, Bowtell D, Brady M, Casado A, Cervantes A, Eisenhauer E, Friedlaender M, Fujiwara K, Grenman S, Guastalla JP, Harper P, Hogberg T, Kaye S, Kitchener H, Kristensen G, Mannel R, Meier W, Miller B, Neijt JP, Oza A, Ozols R, Parmar M, Pecorelli S, Pfisterer J, Poveda A, Provencher D, Pujade-Lauraine E, Randall M, Rochon J, Rustin G, Sagae S, Stehman F, Stuart G, Trimble E, Vasey P, Vergote I, Verheijen R, Wagner U. 2004 consensus statements on the management of ovarian cancer: final document of the 3rd International Gynecologic Cancer Intergroup Ovarian Cancer Consensus Conference (GCIG OCCC 2004) Ann Oncol. 2005;16:viii7–12. doi: 10.1093/annonc/mdi961. - DOI - PubMed
- Ozols RF, Bundy BN, Fowler J, Clarke-Pearson D, Mannel R, Hartenback E, Baergen R. Randomized phase III study of cisplatin (CIS)/paclitaxel (PAC) versus carboplatin (CARBO)/PAC in optimal stage III epithelial ovarian cancer (OC): a Gynecologic Oncology Group trial (GOG 158) Proc Am Soc Clin Oncol. 1999;18:356a.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical