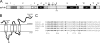Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes - PubMed (original) (raw)
Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes
Clara C Posthuma et al. J Virol. 2008 May.
Abstract
The replication/transcription complex of the arterivirus equine arteritis virus (EAV) is associated with paired membranes and/or double-membrane vesicles (DMVs) that are thought to originate from the endoplasmic reticulum. Previously, coexpression of two putative transmembrane nonstructural proteins (nsp2 and nsp3) was found to suffice to induce these remarkable membrane structures, which are typical of arterivirus infection. Here, site-directed mutagenesis was used to investigate the role of nsp3 in more detail. Liberation of the hydrophobic N terminus of nsp3, which is normally achieved by cleavage of the nsp2/3 junction by the nsp2 protease, was nonessential for the formation of DMVs. However, the substitution of each of a cluster of four conserved cysteine residues, residing in a predicted luminal loop of nsp3, completely blocked DMV formation. Some of these mutant nsp3 proteins were also found to be highly cytotoxic, in particular, exerting a dramatic effect on the endoplasmic reticulum. The functionality of an engineered N glycosylation site in the cysteine-containing loop confirmed both its presence in the lumen and the transmembrane nature of nsp3. This mutant displayed an interesting intermediate phenotype in terms of DMV formation, with paired and curved membranes being formed, but DMV formation apparently being impaired. The effect of nsp3 mutations on replicase polyprotein processing was investigated, and several mutations were found to influence processing of the region downstream of nsp3 by the nsp4 main protease. When tested in an EAV reverse genetics system, none of the nsp3 mutations was tolerated, again underlining the crucial role of the protein in the arterivirus life cycle.
Figures
FIG. 1.
(A) The domain organization of EAV replicase pp1ab. The border between ORF1a- and ORF1b-encoded residues is indicated as the RFS (ribosomal frameshift). Gray and black arrowheads represent the sites that are cleaved by accessory proteases (nsp1 papain-like cysteine protease [PCP] and nsp2 CP) and the nsp4 main protease (MP), respectively. Cleavage products are numbered, and the locations of domains that are structurally or functionally related to those in other nidovirus replicases are highlighted (12). These include four nidovirus-wide conserved domains encoded in ORF1b (RdRp, zinc-binding domain [Z], helicase [HEL], and NendoU [N]) and three putative transmembrane domains (TM1, TM2, and TM3). The α and β symbols refer to the products of the recently documented internal cleavage of nsp7 by the nsp4 protease (44). (B) Predicted membrane topology of EAV nsp3. The four transmembrane helices predicted by ConPred II (
http://bioinfo.si.hirosaki-u.ac.jp/∼ConPred2
) are depicted. Open circles mark the positions of the four conserved Cys residues (see panel C) in the first (predicted) luminal loop. The position of the N glycosylation site engineered at position 873 in the Thr-873→Asn mutation is also indicated. The nsp2/3 and nsp3/4 cleavage sites are indicated by arrowheads and amino acid numbers refer to those in the full-length EAV pp1ab. (C) Arterivirus-wide sequence alignment of the (predicted) luminal loop of nsp3, indicating the four conserved Cys residues (bold) and engineered N glycosylation site (see above). Fully conserved residues are highlighted in bold. LDV-P, lactate dehydrogenase-elevating virus Plagemann strain (accession no. U15146); PRRSV-EU, porcine reproductive and respiratory syndrome virus, strain Lelystad (accession no. M96262); PRRSV-NA, porcine reproductive and respiratory syndrome virus, strain VR2332 (accession no. DQ217415); SHFV, simian hemorrhagic fever virus (accession no. NC_003092); EAV, equine arteritis virus (accession no. NC_002532.).
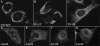
FIG. 2.
Immunofluorescence microscopy analysis of BHK-21 cells transfected with SinRep-5 vectors expressing wild-type and mutant versions of an HA epitope-tagged EAV nsp2-3 polyprotein. Cells were fixed at 8 h posttransfection and stained for nsp2 with a HA tag-specific antibody. Perinuclear staining was observed for the wt nsp2-3 (A) as well as for the polyprotein containing the nsp2 CP-inactivating mutation His-332→Tyr (B). For the polyprotein carrying the Thr-873→Asn N glycosylation mutation in the nsp3 loop, an “intermediate phenotype” was observed, with perinuclear staining and label spread through the cell (C). Expression of nsp2-3 polyproteins carrying mutations in the conserved Cys cluster in nsp3 (D to G) resulted in a dispersed labeling of the cytoplasm and the formation of many vesicles and vacuoles.
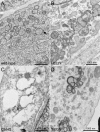
FIG. 3.
Electron micrographs of ultrathin sections of BHK-21 cells transfected with SinRep-5 vectors expressing wild-type and mutant versions of an HA-epitope-tagged EAV nsp2-3 polyprotein (8 h posttransfection). Bars, 1 μm. (A) Formation of DMVs upon expression of the wt nsp2-3 polyprotein. The arrow points to a neck-like connection to an ER cisterna, very similar to those observed previously (37). (B) Cells expressing of a nsp2-3 polyprotein containing an nsp2 CP active site mutation (His-332→Tyr) show normal DMV formation. (C) Example of a cell expressing a nsp2-3 polyprotein carrying a substitution of one of the conserved Cys residues in the nsp3 loop (Cys-864→Ser), which leads to the formation of single-membrane vesicles and large vacuoles that are probably derived from the ER. (D) Expression of the nsp2-3 polyprotein carrying the Thr-873→Asn N glycosylation mutation in the nsp3 loop, which reduced DMV formation and produced conspicuous “balloon-shaped” structures consisting of double-membrane sheets.
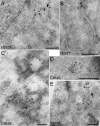
FIG. 4.
Selected close-up electron micrographs of BHK-21 cells expressing mutant EAV nsp2-3 polyproteins. For details, see the legend for Fig. 3. Bars, 200 nm. (A) Typical DMV formed in cells expressing the nsp2-3 polyprotein containing an inactivated nsp2 CP. (B) Example of a single-membrane vesicle induced upon expression of a nsp2-3 polyprotein carrying a Cys-864→Ser substitution in the nsp3 loop. (C) Close-up of “balloon-shaped” structures of paired membranes induced in cells expressing the Thr-873→Asn N glycosylation mutant of nsp2-3.
FIG. 5.
Cryoimmuno-EM images of BHK-21 cells transfected with SinRep-5 vectors expressing mutant HA-tagged EAV nsp2-3 polyproteins. Bars, 200 nm. (A) The nsp2 CP active site mutant (H332Y), for which the nsp2-3 polyprotein remains uncleaved, produced paired membranes (arrow) which double labeled for nsp2 (anti-HA monoclonal antibody; 5 nm gold [open arrowheads]) and nsp3 (anti-nsp3 rabbit antiserum; 10 nm gold [filled arrowheads]). (B) Example of a double-membrane cisterna in cells expressing the H332Y mutant nsp2-3. The nonpaired part of the membranes is double labeled for PDI (anti-PDI monoclonal antibody; 5 nm gold [open arrowheads]) and nsp3 (anti-nsp3 rabbit antiserum; 10 nm gold [filled arrowheads]). (C) Cells expressing nsp2-3 with a Cys-864→Ser replacement in the nsp3 loop. DMVs could not be detected and the labeling for nsp2 and nsp3 was localized to vesicular structures of variable size that were bounded by a single membrane (nsp2 labeled with anti-HA monoclonal antibody, 5 nm gold [open arrowheads], and nsp3 labeled with anti-nsp3 rabbit antiserum, 10 nm gold [filled arrowheads]). (D) Example of membrane structures in cells expressing the C864S mutant nsp2-3 that were double labeled for PDI (anti-PDI monoclonal antibody; 5 nm gold [open arrowheads]) and nsp3 (anti-nsp3 rabbit antiserum; 10 nm gold [filled arrowheads]). (E) Cells expressing nsp2-3 containing the Thr-873→Asn glycosylation mutation in the nsp3 loop. This mutant can produce paired membranes (arrow) and DMVs similar to those seen for the wild-type control, with both structures double labeling for nsp2 (anti-HA monoclonal antibody; 5 nm gold [open arrowheads]) and nsp3 (anti-nsp3 rabbit antiserum; 10 nm gold [filled arrowheads]). However, there are also aggregates of label elsewhere in the cytoplasm (boxed in white), possibly associated with additional membranes structures.
FIG. 6.
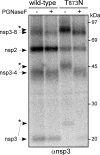
Analysis of cleavage products of wt EAV pp1a and a mutant pp1a carrying the Thr-873→Asn N glycosylation-inducing mutation, which was produced using the recombinant vaccinia virus-driven T7 expression system. Following 35S labeling and immunoprecipitation with an nsp3 antiserum, half of the precipitated proteins were treated with PGNaseF to remove N-linked sugars and proteins were separated by using a 10% SDS-PAGE gel. For nsp3 and the nsp3-4 and nsp3-8 intermediates of the T873N mutant, a mobility shift (bands labeled with asterisks) could be observed; the shift was reversed after the removal of N-linked glycans by PGNaseF. −, absence of; +, presence of.
FIG. 7.
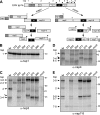
The effect of nsp3 mutations on proteolytic processing of EAV pp1a. Replicase pp1a was expressed from vector pL1a using the recombinant vaccinia virus-driven T7 expression system and RK13 cells. Following 35S labeling, cleavage products were immunoprecipitated using a panel of antisera and separated in a 10% SDS-PAGE gel. (A) Schematic overview of the major and minor pp1a processing pathways of EAV pp1a (51). The interaction of cleaved nsp2 with nsp3 is a trigger for processing via the major pathway, which is initiated by cleavage of the nsp4/5 site. PCP, papain-like cysteine protease; MP, main protease. (B) Immunoprecipitation of nsp1, which was used to establish that the transfection efficiency was similar in all samples. (C to E) Analysis of pp1a cleavage products after immunoprecipitation with nsp3 (C), nsp4 (D), or nsp7-8 (E) antisera (α-nsp). Indicated are the positions of the cleavage products, the size markers, and a novel cleavage product (arrowhead) produced by the four nsp3 Cys mutants, which was tentatively identified as nsp3-5 (see text). Bands marked with asterisks indicate products with altered mobility due to the presence of the Thr-873→Asn glycosylation mutation in nsp3.
Similar articles
- Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication.
Oudshoorn D, Rijs K, Limpens RWAL, Groen K, Koster AJ, Snijder EJ, Kikkert M, Bárcena M. Oudshoorn D, et al. mBio. 2017 Nov 21;8(6):e01658-17. doi: 10.1128/mBio.01658-17. mBio. 2017. PMID: 29162711 Free PMC article. - Biogenesis and architecture of arterivirus replication organelles.
van der Hoeven B, Oudshoorn D, Koster AJ, Snijder EJ, Kikkert M, Bárcena M. van der Hoeven B, et al. Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review. - Antiviral Innate Immune Response Interferes with the Formation of Replication-Associated Membrane Structures Induced by a Positive-Strand RNA Virus.
Oudshoorn D, van der Hoeven B, Limpens RW, Beugeling C, Snijder EJ, Bárcena M, Kikkert M. Oudshoorn D, et al. mBio. 2016 Dec 6;7(6):e01991-16. doi: 10.1128/mBio.01991-16. mBio. 2016. PMID: 27923923 Free PMC article. - Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex.
Snijder EJ, van Tol H, Roos N, Pedersen KW. Snijder EJ, et al. J Gen Virol. 2001 May;82(Pt 5):985-994. doi: 10.1099/0022-1317-82-5-985. J Gen Virol. 2001. PMID: 11297673 - The cell edit: Looking at and beyond non-structural proteins to understand membrane rearrangement in coronaviruses.
Denker L, Dixon AM. Denker L, et al. Arch Biochem Biophys. 2024 Feb;752:109856. doi: 10.1016/j.abb.2023.109856. Epub 2023 Dec 15. Arch Biochem Biophys. 2024. PMID: 38104958 Review.
Cited by
- Mechanism, structural and functional insights into nidovirus-induced double-membrane vesicles.
Wang X, Chen Y, Qi C, Li F, Zhang Y, Zhou J, Wu H, Zhang T, Qi A, Ouyang H, Xie Z, Pang D. Wang X, et al. Front Immunol. 2024 Jun 11;15:1340332. doi: 10.3389/fimmu.2024.1340332. eCollection 2024. Front Immunol. 2024. PMID: 38919631 Free PMC article. Review. - Oligomeric assembly of the C-terminal and transmembrane region of SARS-CoV-2 nsp3.
Babot M, Boulard Y, Agouda S, Pieri L, Fieulaine S, Bressanelli S, Gervais V. Babot M, et al. J Virol. 2024 Apr 16;98(4):e0157523. doi: 10.1128/jvi.01575-23. Epub 2024 Mar 14. J Virol. 2024. PMID: 38483167 Free PMC article. - Classification, replication, and transcription of Nidovirales.
Liao Y, Wang H, Liao H, Sun Y, Tan L, Song C, Qiu X, Ding C. Liao Y, et al. Front Microbiol. 2024 Jan 24;14:1291761. doi: 10.3389/fmicb.2023.1291761. eCollection 2023. Front Microbiol. 2024. PMID: 38328580 Free PMC article. Review. - Research Progress of Porcine Reproductive and Respiratory Syndrome Virus NSP2 Protein.
Liu B, Luo L, Shi Z, Ju H, Yu L, Li G, Cui J. Liu B, et al. Viruses. 2023 Nov 24;15(12):2310. doi: 10.3390/v15122310. Viruses. 2023. PMID: 38140551 Free PMC article. Review. - The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order.
Zhou Z, Qiu Y, Ge X. Zhou Z, et al. Anim Dis. 2021;1(1):5. doi: 10.1186/s44149-021-00005-9. Epub 2021 Apr 23. Anim Dis. 2021. PMID: 34778878 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources