Towards a molecular understanding of symbiont function: identification of a fungal gene for the degradation of xylan in the fungus gardens of leaf-cutting ants - PubMed (original) (raw)
Towards a molecular understanding of symbiont function: identification of a fungal gene for the degradation of xylan in the fungus gardens of leaf-cutting ants
Morten Schiøtt et al. BMC Microbiol. 2008.
Abstract
Background: Leaf-cutting ants live in symbiosis with a fungus that they rear for food by providing it with live plant material. Until recently the fungus' main inferred function was to make otherwise inaccessible cell wall degradation products available to the ants, but new studies have shed doubt on this idea. To provide evidence for the cell wall degrading capacity of the attine ant symbiont, we designed PCR primers from conserved regions of known xylanase genes, to be used in PCR with genomic DNA from the symbiont as template. We also measured xylanase, cellulase and proteinase activities in the fungus gardens in order to investigate the dynamics of degradation activities.
Results: We cloned a xylanase gene from the mutualistic fungus of Acromyrmex echinatior, determined its protein sequence, and inserted it in a yeast expression vector to confirm its substrate specificity. Our results show that the fungus has a functional xylanase gene. We also show by lab experiments in vivo that the activity of fungal xylanase and cellulase is not evenly distributed, but concentrated in the lower layer of fungus gardens, with only modest activity in the middle layer where gongylidia are produced and intermediate activity in the newly established top layer. This vertical distribution appears to be negatively correlated with the concentration of glucose, which indicates a directly regulating role of glucose, as has been found in other fungi and has been previously suggested for the ant fungal symbiont.
Conclusion: The mutualistic fungus of Acromyrmex echinatior has a functional xylanase gene and is thus presumably able to at least partially degrade the cell walls of leaves. This finding supports a saprotrophic origin of the fungal symbiont. The observed distribution of enzyme activity leads us to propose that leaf-substrate degradation in fungus gardens is a multi-step process comparable to normal biodegradation of organic matter in soil ecosystems, but with the crucial difference that a single fungal symbiont realizes most of the steps that are normally provided by a series of microorganisms that colonize fallen leaves in a distinct succession.
Figures
Figure 1
Plant cell wall degradation. Schematic overview of the structure of a plant cell wall and the most important enzymatic reactions involved in the degradation of its polysaccharides. Cellulose microfibrils (blue) are cross-linked by hemicellulose chains (black) within a matrix of pectin (orange). The complex polysaccharides are degraded to disaccharides and oligosaccharides, which are further degraded to soluble monosaccharides that can be assimilated. Full degradation of hemicellulose and pectin may involve more enzymes than those presented here (up to ca. 17 for hemicellulose and ca. 24 for pectin [23]).
Figure 2
Xylanase sequence. cDNA and protein sequence of the LgXyn1 xylanase gene in the Leucoagaricus gongylophorus fungal symbiont of the leaf-cutting ant Acromyrmex echinatior. Numbering of amino acids and nucleotides starts from the start codon (ATG). The position of the single intron of this gene is indicated by an arrow, and the stop codon is indicated by an asterisk.
Figure 3
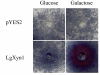
Heterologous expression of xylanase gene. Activity plate assays with Azur-linked Xylan (AZCL, Megazyme). The two top panels show assays using extracts of yeast cells transformed with an empty vector (pYES2), which was expected to give no reaction. The two bottom panels show assays using extracts of yeast cells transformed with cDNA of LgXyn1 inserted in pYES2. These show a clear indication of xylanase activity, with the strongest activity being observed when the yeast cells were grown on galactose-containing media, which induces transcription of the inserted cDNA.
Figure 4
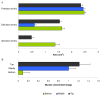
Enzyme activities and glucose concentration infungus gardens. A, enzyme activities in samples of fungus garden material from the top, middle or bottom layer o f the fungus gardens of three A. echinatior colonies (Ae150, Ae219, Ae322) measured as the area of the blue halo 22 hours after incubation on AZCL-casein, AZCL-HE-cellulose or AZCL-xylan (see B-panel for column identities). B, glucose concentration in the top, middle and bottom layer of the fungus garden of colony Ae322 in mg glucose per g fungus garden. All error bars are SEs.
Similar articles
- Ant mediated redistribution of a xyloglucanase enzyme in fungus gardens of Acromyrmex echinatior.
Kooij PW, Pullens JW, Boomsma JJ, Schiøtt M. Kooij PW, et al. BMC Microbiol. 2016 May 6;16:81. doi: 10.1186/s12866-016-0697-4. BMC Microbiol. 2016. PMID: 27154066 Free PMC article. - The fungal symbiont of Acromyrmex leaf-cutting ants expresses the full spectrum of genes to degrade cellulose and other plant cell wall polysaccharides.
Grell MN, Linde T, Nygaard S, Nielsen KL, Boomsma JJ, Lange L. Grell MN, et al. BMC Genomics. 2013 Dec 28;14:928. doi: 10.1186/1471-2164-14-928. BMC Genomics. 2013. PMID: 24373541 Free PMC article. - Leaf-cutting ant fungi produce cell wall degrading pectinase complexes reminiscent of phytopathogenic fungi.
Schiøtt M, Rogowska-Wrzesinska A, Roepstorff P, Boomsma JJ. Schiøtt M, et al. BMC Biol. 2010 Dec 31;8:156. doi: 10.1186/1741-7007-8-156. BMC Biol. 2010. PMID: 21194476 Free PMC article. - The prominent role of fungi and fungal enzymes in the ant-fungus biomass conversion symbiosis.
Lange L, Grell MN. Lange L, et al. Appl Microbiol Biotechnol. 2014 Jun;98(11):4839-51. doi: 10.1007/s00253-014-5708-5. Epub 2014 Apr 15. Appl Microbiol Biotechnol. 2014. PMID: 24728757 Review. - The origin of the attine ant-fungus mutualism.
Mueller UG, Schultz TR, Currie CR, Adams RM, Malloch D. Mueller UG, et al. Q Rev Biol. 2001 Jun;76(2):169-97. doi: 10.1086/393867. Q Rev Biol. 2001. PMID: 11409051 Review.
Cited by
- Fungal plant cell wall-degrading enzyme database: a platform for comparative and evolutionary genomics in fungi and Oomycetes.
Choi J, Kim KT, Jeon J, Lee YH. Choi J, et al. BMC Genomics. 2013;14 Suppl 5(Suppl 5):S7. doi: 10.1186/1471-2164-14-S5-S7. Epub 2013 Oct 16. BMC Genomics. 2013. PMID: 24564786 Free PMC article. - Leucoagaricus gongylophorus produces diverse enzymes for the degradation of recalcitrant plant polymers in leaf-cutter ant fungus gardens.
Aylward FO, Burnum-Johnson KE, Tringe SG, Teiling C, Tremmel DM, Moeller JA, Scott JJ, Barry KW, Piehowski PD, Nicora CD, Malfatti SA, Monroe ME, Purvine SO, Goodwin LA, Smith RD, Weinstock GM, Gerardo NM, Suen G, Lipton MS, Currie CR. Aylward FO, et al. Appl Environ Microbiol. 2013 Jun;79(12):3770-8. doi: 10.1128/AEM.03833-12. Epub 2013 Apr 12. Appl Environ Microbiol. 2013. PMID: 23584789 Free PMC article. - The Evolutionary Innovation of Nutritional Symbioses in Leaf-Cutter Ants.
Aylward FO, Currie CR, Suen G. Aylward FO, et al. Insects. 2012 Jan 6;3(1):41-61. doi: 10.3390/insects3010041. Insects. 2012. PMID: 26467948 Free PMC article. Review. - Lessons From Insect Fungiculture: From Microbial Ecology to Plastics Degradation.
Barcoto MO, Rodrigues A. Barcoto MO, et al. Front Microbiol. 2022 May 24;13:812143. doi: 10.3389/fmicb.2022.812143. eCollection 2022. Front Microbiol. 2022. PMID: 35685924 Free PMC article. - Battle through signaling between wheat and the fungal pathogen Septoria tritici revealed by proteomics and phosphoproteomics.
Yang F, Melo-Braga MN, Larsen MR, Jørgensen HJ, Palmisano G. Yang F, et al. Mol Cell Proteomics. 2013 Sep;12(9):2497-508. doi: 10.1074/mcp.M113.027532. Epub 2013 May 29. Mol Cell Proteomics. 2013. PMID: 23722186 Free PMC article.
References
- Weber NA. Gardening Ants: The Attines. Philadelphia: American Philosophical Society; 1972.
- Cherrett JM, Powell R, Stradling DJ. The mutualism between leaf-cutting ants and their fungus. In: Wilding N, Collins NM, Webber JF, editor. Insect-Fungus Interactions. London: Academic Press; 1989. pp. 93–120.
- Hölldobler B, Wilson EO. The Ants. Cambridge: Harvard University Press; 1990.
- Abril AB, Bucher EH. Evidence that the fungus cultured by leaf-cutting ants does not metabolize cellulose. Ecol Lett. 2002;5:325–328. doi: 10.1046/j.1461-0248.2002.00327.x. - DOI
- Abril AB, Bucher EH. Nutritional sources of the fungus cultured by leaf-cutting ants. Appl Soil Ecol. 2004;26:243–247. doi: 10.1016/j.apsoil.2003.12.008. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous