The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues - PubMed (original) (raw)
The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues
Andrea Scrima et al. EMBO J. 2008.
Abstract
The GTP-binding protein Rap1 regulates integrin-mediated and other cell adhesion processes. Unlike most other Ras-related proteins, it contains a threonine in switch II instead of a glutamine (Gln61 in Ras), a residue crucial for the GTPase reaction of most G proteins. Furthermore, unlike most other GTPase-activating proteins (GAPs) for small G proteins, which supply a catalytically important Arg-finger, no arginine residue of RapGAP makes a significant contribution to the GTPase reaction of Rap1. For a detailed understanding of the reaction mechanism, we have solved the structure of Rap1 in complex with Rap1GAP. It shows that the Thr61 of Rap is away from the active site and that an invariant asparagine of RapGAPs, the Asn-thumb, takes over the role of the cis-glutamine of Ras, Rho or Ran. The structure and biochemical data allow to further explain the mechanism and to define the important role of a conserved tyrosine. The structure and biochemical data furthermore show that the RapGAP homologous region of the tumour suppressor Tuberin is sufficient for catalysis on Rheb.
Figures
Figure 1
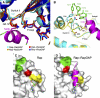
Rap–Rap1GAP complex and interface analysis. (A) Ribbon representation of the Rap–GDP·BeF3−-Rap1GAP complex with Rap1B in cyan and Rap1GAP in green (catalytic domain green; dimerisation domain olive green). The catalytic helix containing the Asn-thumb (Asn290) is shown in magenta, GDP-BeF3− as ball-and-stick. (B) Schematic representation of interacting residues. Interactions shown in detail in (C–E) are depicted with a dashed line. (C–E) Structural details of interactions between Rap1B and Rap1GAP, with colours as in (A). (F) HPLC-based analysis of the Rap-stimulated GTPase reaction, with 200 μM wt and mutant Rap and 100 nM Rap1GAP. (G) Stopped-flow analysis of the interaction between 2 μM Aedans-labelled wt and mutant Rap and 50 μM Rap1GAP; reaction was followed by monitoring fluorescence through a 408 nm cutoff filter. Wt and mutant Rap contain the A86C mutation, which has been shown to behave as wild type, as described earlier (Kraemer et al, 2002; Chakrabarti et al, 2007).
Figure 2
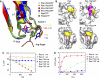
The active site. (A) Active site of Rap–Rap1GAP, shown as superimposition with Ras, Ran and Rho in complex with their cognate GAPs. Asn290 in Rap1GAP occupies the position of the catalytic Gln in Ras (Gln61), Ran (Gln69) and Rho (Gln63). The G12 position in Rap1B is marked with a sphere. (B) Superimposition of uncomplexed Rap (yellow) and the Rap–Rap1GAP (cyan/green) complex. Interaction of Gln63 and Phe64 with residues on Rap1GAP (green) forces switch II into an alternative conformation (arrows) to release blockade by Thr61 thereby allowing Asn290 to enter the active site. (C, D) Surface representation of uncomplexed Rap (C) and Rap in complex with Rap1GAP (D). The switch II residues T61 (red) and Q63/F64 (green) undergo a drastic conformational change upon complex formation with Rap1GAP to allow access for the Asn-thumb to the active site.
Figure 3
Role of Tyr32. (A) Superimposition of active sites from various structures as indicated, with an emphasis on the conformation of Tyr32. Systems using an Arg-finger (RasGAP/RhoGAP) show Tyr32 in a more open versus a more closed conformation for Ran and Rap. (B) Surface representation of uncomplexed Rap/Ras or in complex with their respective GAPs, with residue Tyr32 labelled in yellow. The Rap or Ras structures are shown in slightly different orientations. Catalytic elements from the GAP, the catalytic helix with the Asn-thumb and the Arg-finger (magenta and brown, respectively) are shown as ribbon. (C) HPLC-based analysis of the GTPase stimulation by Rap1GAP for Rap wt and Y32-mutants (described in Figure 1F). (D) GTPase stimulation of Rap wt and Y32 mutants analysed by radioactive charcoal assay. Rap (10 μM) and Rap1GAP (50 nM) were incubated as described before (Kupzig et al, 2006). The concentration of released 32P-labelled Pi (correscponding to hydrolysed GTP) was plotted against the reaction time and initial rates were determined by linear regression fitting.
Figure 4
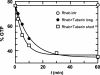
Stimulation of Rheb GTP hydrolysis by Tuberin. HPLC-based analysis of intrinsic and Tuberin-stimulated GTPase of Rheb, with two different constructs of the catalytic domain of Tuberin, using 80 μM Rheb and 100 μM Tuberin (Tuberinlong: 1532–1760; Tuberinshort: 1538–1729).
Similar articles
- The GTPase-activating protein Rap1GAP uses a catalytic asparagine.
Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A. Daumke O, et al. Nature. 2004 May 13;429(6988):197-201. doi: 10.1038/nature02505. Nature. 2004. PMID: 15141215 - Insight into catalysis of a unique GTPase reaction by a combined biochemical and FTIR approach.
Chakrabarti PP, Daumke O, Suveyzdis Y, Kötting C, Gerwert K, Wittinghofer A. Chakrabarti PP, et al. J Mol Biol. 2007 Apr 6;367(4):983-95. doi: 10.1016/j.jmb.2006.11.022. Epub 2006 Nov 10. J Mol Biol. 2007. PMID: 17300802 - Structural basis for the unique biological function of small GTPase RHEB.
Yu Y, Li S, Xu X, Li Y, Guan K, Arnold E, Ding J. Yu Y, et al. J Biol Chem. 2005 Apr 29;280(17):17093-100. doi: 10.1074/jbc.M501253200. Epub 2005 Feb 23. J Biol Chem. 2005. PMID: 15728574 - RapGAPs in brain: multipurpose players in neuronal Rap signalling.
Spilker C, Kreutz MR. Spilker C, et al. Eur J Neurosci. 2010 Jul;32(1):1-9. doi: 10.1111/j.1460-9568.2010.07273.x. Epub 2010 Jun 22. Eur J Neurosci. 2010. PMID: 20576033 Review. - Regulating Rap small G-proteins in time and space.
Gloerich M, Bos JL. Gloerich M, et al. Trends Cell Biol. 2011 Oct;21(10):615-23. doi: 10.1016/j.tcb.2011.07.001. Epub 2011 Aug 4. Trends Cell Biol. 2011. PMID: 21820312 Review.
Cited by
- Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases.
Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, Nakayama E, Okawa K, Nureki O, Kimura T, Kita T, Horiuchi H. Shirakawa R, et al. J Biol Chem. 2009 Aug 7;284(32):21580-8. doi: 10.1074/jbc.M109.012112. Epub 2009 Jun 11. J Biol Chem. 2009. PMID: 19520869 Free PMC article. - It takes two to tango: regulation of G proteins by dimerization.
Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. Gasper R, et al. Nat Rev Mol Cell Biol. 2009 Jun;10(6):423-9. doi: 10.1038/nrm2689. Epub 2009 May 8. Nat Rev Mol Cell Biol. 2009. PMID: 19424291 Review. - Autoinhibition and signaling by the switch II motif in the G-protein chaperone of a radical B12 enzyme.
Lofgren M, Koutmos M, Banerjee R. Lofgren M, et al. J Biol Chem. 2013 Oct 25;288(43):30980-9. doi: 10.1074/jbc.M113.499970. Epub 2013 Aug 30. J Biol Chem. 2013. PMID: 23996001 Free PMC article. - Case Report: A Novel Missense Variant in the SIPA1L3 Gene Associated With Cataracts in a Chinese Family.
Yang D, Zhou H, Lin J, Zhao S, Zhou H, Yin Z, Ni B, Chen Y, Xie W. Yang D, et al. Front Genet. 2021 Sep 16;12:715599. doi: 10.3389/fgene.2021.715599. eCollection 2021. Front Genet. 2021. PMID: 34603379 Free PMC article. - Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization.
Wang Y, He H, Srivastava N, Vikarunnessa S, Chen YB, Jiang J, Cowan CW, Zhang X. Wang Y, et al. Sci Signal. 2012 Jan 17;5(207):ra6. doi: 10.1126/scisignal.2002636. Sci Signal. 2012. PMID: 22253263 Free PMC article.
References
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 - PubMed
- Bi X, Corpina RA, Goldberg J (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419: 271–277 - PubMed
- Bos JL, de Rooij J, Reedquist KA (2001) Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol 2: 369–377 - PubMed
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129: 865–877 - PubMed
- Brinkmann T, Daumke O, Herbrand U, Kühlmann D, Stege P, Ahmadian MR, Wittinghofer A (2002) Rap-specific GTPase activating protein follows an alternative mechanism. J Biol Chem 277: 12525–12531 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous