Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism - PubMed (original) (raw)
Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism
Daisy Lin et al. Mol Cell Biol. 2008 May.
Abstract
Translational repressors, increasing evidence suggests, participate in the regulation of protein synthesis at the synapse, thus providing a basis for the long-term plastic modulation of synaptic strength. Dendritic BC1 RNA is a non-protein-coding RNA that represses translation at the level of initiation. However, the molecular mechanism of BC1 repression has remained unknown. Here we identify the catalytic activity of eukaryotic initiation factor 4A (eIF4A), an ATP-dependent RNA helicase, as a target of BC1-mediated translational control. BC1 RNA specifically blocks the RNA duplex unwinding activity of eIF4A but, at the same time, stimulates its ATPase activity. BC200 RNA, the primate-specific BC1 counterpart, targets eIF4A activity in identical fashion, as a result decoupling ATP hydrolysis from RNA duplex unwinding. In vivo, BC1 RNA represses translation of a reporter mRNA with 5' secondary structure. The eIF4A mechanism places BC RNAs in a central position to modulate protein synthesis in neurons.
Figures
FIG. 1.
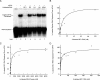
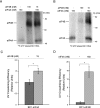
BC1 RNA binds to eIF4A with high affinity. Heterologous and homologous competitive binding assays were performed with labeled and unlabeled RNAs as indicated, and complexes were resolved by EMSA. (A) BC1 RNA specifically displaced RNA duplexes from eIF4A (lanes 2 and 3), while U6 snRNA, tRNA, and an irrelevant RNA (random-sequence [RS] RNA) did not. Also, U4 snRNA, although slightly more substrate competitive, did not effectively displace duplex RNA from eIF4A. Longer exposure of gels (not shown) revealed residual duplex-eIF4A complexes at decreasing levels even in the presence of >10 nM BC1 RNA. The presence (+) or absence (−) of eIF4A and unlabeled RNA is shown above the gel. (B) Using the EMSA approach, complexes of eIF4A with duplex RNA substrate were titrated with BC1 RNA. Nonlinear regression revealed that BC1 RNA displaced RNA duplexes from eIF4A with an apparent IC50 of 8.5 nM. (C) In converse titrations under identical conditions, an apparent IC50 of 308 nM was obtained for the displacement of BC1 RNA from eIF4A by duplex RNA substrate. (D) In homologous competitive binding experiments, complexes of eIF4A with labeled BC1 RNA were titrated with unlabeled BC1 RNA. Nonlinear regression revealed an IC50 of 10 nM, on the basis of which an equilibrium dissociation constant of 0.2 nM was calculated (37) for the binding affinity between BC1 RNA and eIF4A.
FIG. 2.
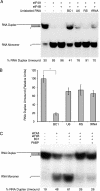
BC1 RNA inhibits eIF4A helicase activity. RNA duplexes (10/44 nt, radiolabeled on the shorter strand) were used as substrates for eIF4A-dependent helicase activity in the presence of 1 mM ATP. (A) BC1 RNA specifically inhibited eIF4A-dependent RNA duplex unwinding. Other small untranslated RNAs, such as U6 snRNA, irrelevant random-sequence (RS) RNA, and a tRNA, did not significantly inhibit unwinding. The presence (+) or absence (−) of eIF4A, eIF4B, and unlabeled RNA is shown above the gel. The percentage of RNA duplex unwound for each reaction mixture represents the ratio of monomer signal divided by the sum of monomer signal plus duplex signal (as determined by phosphorimaging) and is shown below the gel. (B) Quantitative analysis confirmed that BC1 RNA specifically repressed eIF4A-dependent RNA unwinding. Results shown are corrected for spontaneous unwinding (i.e., dissociation into monomers in the absence of eIFs 4A and 4B). Data are presented in the format mean ± standard error of the mean (error bar) (n = 4). For statistical analysis, the Kruskal-Wallis nonparametric one-way analysis of variance was used. A P of <0.05 was obtained and considered significant. For post hoc analysis, the Mann-Whitney U test (comparison with reaction mixtures with no unlabeled RNA) was used, and the value for the reaction mixture with no unlabeled RNA was significantly different (P < 0.05) from the value for the reaction mixture containing BC1 RNA, as indicated by a bracket and asterisk. (C) PABP did not alter BC1-mediated inhibition of eIF4A-dependent and eIF4B-stimulated helicase activity. The duplex RNA substrate used in panel C was generated at a 1:1.5 strand ratio (see Materials and Methods) and displayed a higher overall degree of annealing than the one used in panel A (1:1.25 ratio). The resulting overall lower unwinding efficiency likely explains a correspondingly stronger dependence on eIF4B.
FIG. 3.
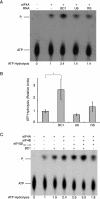
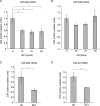
BC1 RNA stimulates eIF4A ATPase activity. Products of [γ-32P]ATP hydrolysis were resolved by TLC. (A) BC1 RNA specifically stimulated eIF4A-mediated ATP hydrolysis, whereas U6 RNA and an irrelevant random-sequence (RS) RNA did not. The presence (+) or absence (−) of eIF4A and RNA is shown above the gel. The percentage of ATP hydrolysis for each reaction mixture was calculated as the ratio between Pi band intensity and the sum of Pi and ATP band intensities and is shown below the gel. (B) Quantitative analysis of ATP hydrolysis. For statistical analysis (n = 4), the Kruskal-Wallis test was used (P < 0.05). The Mann-Whitney U test was used to compare reaction mixtures with no RNA. The value for reaction mixture with no RNA was significantly different (P < 0.05) from the value for BC1 RNA, as indicated by a bracket and asterisk. (C) In the presence of eIF4B and an eIF4A-binding central domain of eIF4G (eIF4GI737-1116), the basal ATPase activity of eIF4A was moderately elevated, and this elevated level was analogously BC1 stimulated about threefold.
FIG. 4.
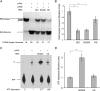
BC200 RNA targets eIF4A catalytic activity in a manner indistinguishable from that of BC1 RNA. Helicase and ATPase assays were performed as described in the legends to Fig. 2 and 3. (A and B) BC200 RNA inhibited eIF4A helicase activity. For statistical analysis (n = 4), the Kruskal-Wallis test was used (P < 0.01). The Mann-Whitney U test was used to compare reaction mixtures with no unlabeled RNA. The value for the reaction mixture with no unlabeled RNA was significantly different (P < 0.05) from the values for the reaction mixtures containing BC1 RNA and BC200 RNA, as indicated by brackets and asterisks. (C and D) BC200 RNA stimulated eIF4A ATPase activity. For statistical analysis (n = 4), the Kruskal-Wallis test was used (P < 0.05). The Mann-Whitney U test was used to compare reaction mixtures with no RNA. The value for the reaction mixture with no RNA was significantly different (P < 0.05) from the value for the reaction mixture with BC200 RNA, as indicated by a bracket and asterisk. BC200 RNA and U6 RNA were used at 10 μM.
FIG. 5.
Recruitment of eIFs 4A and 4B to BC1 RNA is mutually synergistic. UV cross-linking assays were performed with BC1 RNA labeled by in vitro transcription in the presence of either [32P]UTP (A) or [32P]ATP (B). (A and B) If offered jointly, eIFs 4A and 4B mutually stimulated each other's binding interactions with BC1 RNA. Note that eIF4B was offered at a 10-fold-lower concentration than eIF4A. Experiments were performed in the presence of 5 mM ATP. (C and D) Quantitative analysis of UV cross-linking. (C) eIF4B significantly enhanced BC1-eIF4A UV cross-linking efficiency as evaluated by a nonparametric Mann-Whitney U test (compared with reaction mixtures with no [−] eIF4B) (P < 0.05) (n = 4), as is indicated by a bracket and asterisk. (D) eIF4A significantly enhanced BC1-eIF4B UV cross-linking efficiency, as evaluated by a nonparametric Mann-Whitney U test (compared with reaction mixtures with no eIF4A) (P < 0.05) (n = 4) and indicated by a bracket and asterisk.
FIG. 6.
BC1-eIF4A interactions are modulated by ATP and ADP. (A) ATP increased BC1-eIF4A cross-linking efficiency more than 10-fold (compared with cross-linking in the absence of nucleotide). ADP increased cross-linking efficiency fourfold. BC1 RNA was in vitro transcribed in the presence of 32P-labeled ATP and 4-thio-UTP. The presence (+) or absence (−) of eIF4A and nucleotide is shown above the gel. (B) Quantitative analysis of BC1-eIF4A UV cross-linking efficiency. For statistical analysis (n = 4), the Kruskal-Wallis nonparametric one-way analysis of variance was used (P < 0.05). For post hoc analysis, the Mann-Whitney U test was used to compare reaction mixtures with ATP. The value for reaction mixtures containing ATP was significantly different (P < 0.05) from the values for reaction mixtures with ADP and without nucleotides, as is indicated by brackets and asterisks.
FIG. 7.
BC1 RNA significantly represses translation of CAT mRNA in HEK293 cells. (A and B) BC1 RNA significantly inhibited translation of CAT mRNA (5′ capped and A98 adenylated), while U6 RNA did not. Total amounts of transfected small npcRNAs were kept constant by supplementation with random-sequence (RS) RNA to a combined total of 20 pmol. RS RNA thus served as an internal control and calibrator. For statistical analysis, the Kruskal-Wallis test was used (P < 0.05). For post hoc analysis, the Mann-Whitney U test was used to compare reaction mixtures with no BC1 or U6 RNA. Values were significantly different (P < 0.05) from the value for no BC1 RNA for values of BC1 RNA in the range 10 to 20 pmol, as is indicated by brackets and asterisks. (C and D) BC1 RNA significantly repressed translation of polyadenylated and nonadenylated CAT mRNAs. (C) HEK293 cells were cotransfected with CAT-A98 mRNA and either BC1 RNA or U6 RNA. BC1 RNA significantly reduced translation of CAT-A98 compared with U6 RNA. (D) HEK293 cells were cotransfected with CAT-A0 mRNA (5′ capped but not adenylated) and either BC1 RNA or U6 RNA. BC1 RNA significantly inhibited translation of CAT-A0 mRNA compared with U6 RNA. In general, CAT-A0 mRNA was translated less efficiently than CAT-A98 mRNA as noted before (47). Statistical analysis in panels C and D was done by the Mann-Whitney U test. Values that were significantly different (P < 0.05) from the value for the reaction mixture with U6 RNA are indicated by a bracket and asterisk. For panels A to D, n ≥ 4 for each panel shown.
FIG. 8.
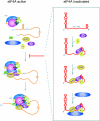
BC1 RNA targets eIF4A functionality. The recruitment of eIFs 4A and 4B to BC1 RNA is mutually synergistic. As a result of BC1-eIF4A interactions, the helicase activity of the factor is uncoupled from its ATPase activity. PABP binds to BC1 RNA concurrently with eIF4A (65, 66) and interacts with eIF4B (10). BC1-eIF4A interactions do not prevent eIF4A from binding to the central domain of eIF4G. PABP is shown interacting with the central A22 domain, eIF4A with the A-rich single-stranded region at the interface to the 3′ terminal stem-loop, and eIF4B with the terminal stem-loop itself. Although speculative, this placement is compatible with the BC1-eIF4A-eIF4B cross-linking data (Fig. 5) (see Fig. S6 in the supplemental material) that suggest that eIF4B is recruited by eIF4A into cross-linking proximity of the A-rich eIF4A-interacting domain. Standard 48S complex formation is shown on the left (see also reference 26). Some initiation factors have been omitted for clarity. The shown BC1 secondary structure sketch is based on previous experimental work (55). The full secondary structure of BC1 RNA is shown in Fig. S10 in the supplemental material.
Similar articles
- The DEAD-box helicase eIF4A: paradigm or the odd one out?
Andreou AZ, Klostermeier D. Andreou AZ, et al. RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review. - Dendritic BC1 RNA in translational control mechanisms.
Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, Hellen CU, Tiedge H. Wang H, et al. J Cell Biol. 2005 Dec 5;171(5):811-21. doi: 10.1083/jcb.200506006. J Cell Biol. 2005. PMID: 16330711 Free PMC article. - Dendritic BC1 RNA: functional role in regulation of translation initiation.
Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Wang H, et al. J Neurosci. 2002 Dec 1;22(23):10232-41. doi: 10.1523/JNEUROSCI.22-23-10232.2002. J Neurosci. 2002. PMID: 12451124 Free PMC article. - The Expression Alteration of BC1 RNA and its Interaction with Eukaryotic Translation Initiation Factor eIF4A Post-Status Epilepticus.
Zeng X, Zong W, Gao Q, Chen S, Chen L, Zeng G, Huang W, Li Z, Zeng C, Xie Y, Li X, Xiao B, Dongsheng-Ouyang, Hu K. Zeng X, et al. Neurochem Res. 2018 Jul;43(7):1328-1338. doi: 10.1007/s11064-018-2548-1. Epub 2018 May 17. Neurochem Res. 2018. PMID: 29774448 Retracted. - Eukaryotic initiation factor 4A (eIF4A) during viral infections.
Montero H, Pérez-Gil G, Sampieri CL. Montero H, et al. Virus Genes. 2019 Jun;55(3):267-273. doi: 10.1007/s11262-019-01641-7. Epub 2019 Feb 22. Virus Genes. 2019. PMID: 30796742 Free PMC article. Review.
Cited by
- The DEAD-box helicase eIF4A: paradigm or the odd one out?
Andreou AZ, Klostermeier D. Andreou AZ, et al. RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review. - Neuronal BC RNA Transport Impairments Caused by Systemic Lupus Erythematosus Autoantibodies.
Muslimov IA, Iacoangeli A, Eom T, Ruiz A, Lee M, Stephenson S, Ginzler EM, Tiedge H. Muslimov IA, et al. J Neurosci. 2019 Sep 25;39(39):7759-7777. doi: 10.1523/JNEUROSCI.1657-18.2019. Epub 2019 Aug 12. J Neurosci. 2019. PMID: 31405929 Free PMC article. - The functional role of long non-coding RNA in human carcinomas.
Gibb EA, Brown CJ, Lam WL. Gibb EA, et al. Mol Cancer. 2011 Apr 13;10:38. doi: 10.1186/1476-4598-10-38. Mol Cancer. 2011. PMID: 21489289 Free PMC article. Review. - Long Non-Coding RNAs in Neuronal Aging.
Pereira Fernandes D, Bitar M, Jacobs FMJ, Barry G. Pereira Fernandes D, et al. Noncoding RNA. 2018 Apr 18;4(2):12. doi: 10.3390/ncrna4020012. Noncoding RNA. 2018. PMID: 29670042 Free PMC article. Review. - The impact of non-coding RNAs: workshop on new functions of regulatory RNAs in pro- & eukaryotes.
Boots JL, Moll I. Boots JL, et al. EMBO Rep. 2009 Jun;10(6):563-7. doi: 10.1038/embor.2009.100. Epub 2009 May 22. EMBO Rep. 2009. PMID: 19465893 Free PMC article. No abstract available.
References
- Abramson, R. D., T. E. Dever, T. G. Lawson, B. K. Ray, R. E. Thach, and W. C. Merrick. 1987. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 2623826-3832. - PubMed
- Barciszewski, J., and V. A. Erdmann (ed.). 2003. Noncoding RNAs: molecular biology and molecular medicine. Landes Bioscience, Georgetown, TX.
- Bleichert, F., and S. J. Baserga. 2007. The long unwinding road of RNA helicases. Mol. Cell 27339-352. - PubMed
- Bordeleau, M. E., R. Cencic, L. Lindqvist, M. Oberer, P. Northcote, G. Wagner, and J. Pelletier. 2006. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 131287-1295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM059660/GM/NIGMS NIH HHS/United States
- R01 AI051340/AI/NIAID NIH HHS/United States
- AI51340/AI/NIAID NIH HHS/United States
- NS046769/NS/NINDS NIH HHS/United States
- R56 AI051340/AI/NIAID NIH HHS/United States
- GM059660/GM/NIGMS NIH HHS/United States
- R01 NS046769/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous