Comprehensive high-throughput arrays for relative methylation (CHARM) - PubMed (original) (raw)
Comprehensive high-throughput arrays for relative methylation (CHARM)
Rafael A Irizarry et al. Genome Res. 2008 May.
Abstract
This study was originally conceived to test in a rigorous way the specificity of three major approaches to high-throughput array-based DNA methylation analysis: (1) MeDIP, or methylated DNA immunoprecipitation, an example of antibody-mediated methyl-specific fractionation; (2) HELP, or HpaII tiny fragment enrichment by ligation-mediated PCR, an example of differential amplification of methylated DNA; and (3) fractionation by McrBC, an enzyme that cuts most methylated DNA. These results were validated using 1466 Illumina methylation probes on the GoldenGate methylation assay and further resolved discrepancies among the methods through quantitative methylation pyrosequencing analysis. While all three methods provide useful information, there were significant limitations to each, specifically bias toward CpG islands in MeDIP, relatively incomplete coverage in HELP, and location imprecision in McrBC. However, we found that with an original array design strategy using tiling arrays and statistical procedures that average information from neighboring genomic locations, much improved specificity and sensitivity could be achieved, e.g., approximately 100% sensitivity at 90% specificity with McrBC. We term this approach "comprehensive high-throughput arrays for relative methylation" (CHARM). While this approach was applied to McrBC analysis, the array design and computational algorithms are fractionation method-independent and make this a simple, general, relatively inexpensive tool suitable for genome-wide analysis, and in which individual samples can be assayed reliably at very high density, allowing locus-level genome-wide epigenetic discrimination of individuals, not just groups of samples. Furthermore, unlike the other approaches, CHARM is highly quantitative, a substantial advantage in application to the study of human disease.
Figures
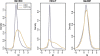
Figure 1.
Density estimates (smoothed histograms) of the _M_-values comparing DKO (gray) and HCT116 (brown) samples. Note that the display of the overall M distribution masks differences at individual sites.
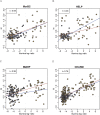
Figure 2.
Comparison of method-specific methylation measurements to reference data. For the HCT116 (brown) and DKO (gray) samples, _M_-values from high-throughput methods are plotted against _M_-values from the Illumina reference platform. To illustrate the CpG observed-to-expected ratio, a 500-bp window was formed around each probe; this ratio (multiplied by 10) is displayed inside each point. A regression line was calculated and is displayed for probes with ratios <0.6 (blue line) and >0.6 (red line).
Figure 3.
DNA fragment-length–related biases. (A) _M_-values for the HCT116 sample are stratified by the DNA fragment size predicted by the McrBC (left panel) and HELP (right panel) enzyme digestions. (B) For all three methods, a 500-bp window was formed around each probe, the observed-to-expected ratio of CpG was calculated, and box-plots of the _M_-values are displayed by these ratios. Only probes related to fragments of sizes between 50 and 600 bp for McrBC, and between 600 and 1200 bp for HELP, are included.
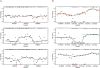
Figure 4.
M values plotted against contiguous locations on the genome for all three methods. The points are the observed _M_-values. The _M_-values for probes in the same predicted segments for McrBC and HELP were averaged and are represented in the figure with orange and green lines, respectively. The data were smoothed using running medians with a window size of 7 and showed the results with black curves. CpG locations are shown as black tick marks at the top of the plots. (A) Segment showing lack of methylation determined by the Illumina platform. (B) Segment with high methylation as determined by the Illumina platform. The Illumina probes and measured methylation percentages are shown on the bottom of the plot.
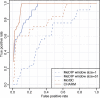
Figure 5.
ROC curve demonstrating the advantage of genome-weighted smoothing. We considered all gene regions represented on the Illumina platform. For the purpose of ROC calculation, highly methylated and unmethylated regions were compared. If all probes in the region showed on the reference Illumina platform a methylation percentage >90%, the region was considered a true positive. If all probes in the region reported a percentage <10% they were considered a true negative. To define a positive from the microarray data using a window size of 1, a cut-off for the _M_-values was chosen. If any probe intensity within the region was above that cut-off, it was defined as positive. A running median with a window size of 51 was then analyzed and defined a positive in the same way, except that the smoothed results instead of the individual probe intensities were used. Results are shown for both McrBC and MeDIP. For a given threshold, the true-positive rate is defined as the percentage of true-positive regions for which the microarray data surpasses that threshold. The false-positive rate is defined in the same way but for the true-negative regions.
Similar articles
- Comprehensive high-throughput arrays for relative methylation (CHARM).
Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Ladd-Acosta C, et al. Curr Protoc Hum Genet. 2010 Apr;Chapter 20:Unit 20.1.1-19. doi: 10.1002/0471142905.hg2001s65. Curr Protoc Hum Genet. 2010. PMID: 20373514 Free PMC article. - Assessment of methylation level prediction accuracy in methyl-DNA immunoprecipitation and sodium bisulfite based microarray platforms.
Rajendram R, Ferreira JC, Grafodatskaya D, Choufani S, Chiang T, Pu S, Butcher DT, Wodak SJ, Weksberg R. Rajendram R, et al. Epigenetics. 2011 Apr;6(4):410-5. doi: 10.4161/epi.6.4.14763. Epub 2011 Apr 1. Epigenetics. 2011. PMID: 21343703 - Systematic evaluation of genome-wide methylated DNA enrichment using a CpG island array.
Yang L, Zhang K, Dai W, He X, Zhao Q, Wang J, Sun ZS. Yang L, et al. BMC Genomics. 2011 Jan 6;12:10. doi: 10.1186/1471-2164-12-10. BMC Genomics. 2011. PMID: 21211017 Free PMC article. - Analyzing the cancer methylome through targeted bisulfite sequencing.
Lee EJ, Luo J, Wilson JM, Shi H. Lee EJ, et al. Cancer Lett. 2013 Nov 1;340(2):171-8. doi: 10.1016/j.canlet.2012.10.040. Epub 2012 Nov 28. Cancer Lett. 2013. PMID: 23200671 Free PMC article. Review. - Genome-scale DNA methylation analysis.
Fouse SD, Nagarajan RO, Costello JF. Fouse SD, et al. Epigenomics. 2010 Feb;2(1):105-17. doi: 10.2217/epi.09.35. Epigenomics. 2010. PMID: 20657796 Free PMC article. Review.
Cited by
- Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu.
Burris HH, Braun JM, Byun HM, Tarantini L, Mercado A, Wright RJ, Schnaas L, Baccarelli AA, Wright RO, Tellez-Rojo MM. Burris HH, et al. Epigenomics. 2013 Jun;5(3):271-81. doi: 10.2217/epi.13.24. Epigenomics. 2013. PMID: 23750643 Free PMC article. - Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host.
Timp W, Feinberg AP. Timp W, et al. Nat Rev Cancer. 2013 Jul;13(7):497-510. doi: 10.1038/nrc3486. Epub 2013 Jun 13. Nat Rev Cancer. 2013. PMID: 23760024 Free PMC article. Review. - Association of the CpG methylation pattern of the proximal insulin gene promoter with type 1 diabetes.
Fradin D, Le Fur S, Mille C, Naoui N, Groves C, Zelenika D, McCarthy MI, Lathrop M, Bougnères P. Fradin D, et al. PLoS One. 2012;7(5):e36278. doi: 10.1371/journal.pone.0036278. Epub 2012 May 2. PLoS One. 2012. PMID: 22567146 Free PMC article. - Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns.
Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, Gingrich JA, Haghighi F, Nutter R, Bestor TH. Edwards JR, et al. Genome Res. 2010 Jul;20(7):972-80. doi: 10.1101/gr.101535.109. Epub 2010 May 20. Genome Res. 2010. PMID: 20488932 Free PMC article. - SPARSE INTEGRATIVE CLUSTERING OF MULTIPLE OMICS DATA SETS.
Shen R, Wang S, Mo Q. Shen R, et al. Ann Appl Stat. 2013 Apr 9;7(1):269-294. doi: 10.1214/12-AOAS578. Ann Appl Stat. 2013. PMID: 24587839 Free PMC article.
References
- Allinen M., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Huang H., Porter D., Hu M., Chin L., Richardson A., Porter D., Hu M., Chin L., Richardson A., Hu M., Chin L., Richardson A., Chin L., Richardson A., Richardson A., et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. - PubMed
- Allison D.B., Cui X., Page G.P., Sabripour M., Cui X., Page G.P., Sabripour M., Page G.P., Sabripour M., Sabripour M. Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. - PubMed
- Bibikova M., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Doucet D., Thomas N.J., Wang Y., Vollmer E., Thomas N.J., Wang Y., Vollmer E., Wang Y., Vollmer E., Vollmer E., et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. - PMC - PubMed
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P., Irizarry R.A., Astrand M., Speed T.P., Astrand M., Speed T.P., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 RR021967/RR/NCRR NIH HHS/United States
- R37 CA054358/CA/NCI NIH HHS/United States
- P50 HG003233/HG/NHGRI NIH HHS/United States
- R01 GM083084/GM/NIGMS NIH HHS/United States
- HG003233/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources