Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways - PubMed (original) (raw)
Comparative Study
Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways
Elodie Gaulin et al. PLoS One. 2008.
Abstract
Aphanomyces euteiches is an oomycete pathogen that causes seedling blight and root rot of legumes, such as alfalfa and pea. The genus Aphanomyces is phylogenically distinct from well-studied oomycetes such as Phytophthora sp., and contains species pathogenic on plants and aquatic animals. To provide the first foray into gene diversity of A. euteiches, two cDNA libraries were constructed using mRNA extracted from mycelium grown in an artificial liquid medium or in contact to plant roots. A unigene set of 7,977 sequences was obtained from 18,864 high-quality expressed sequenced tags (ESTs) and characterized for potential functions. Comparisons with oomycete proteomes revealed major differences between the gene content of A. euteiches and those of Phytophthora species, leading to the identification of biosynthetic pathways absent in Phytophthora, of new putative pathogenicity genes and of expansion of gene families encoding extracellular proteins, notably different classes of proteases. Among the genes specific of A. euteiches are members of a new family of extracellular proteins putatively involved in adhesion, containing up to four protein domains similar to fungal cellulose binding domains. Comparison of A. euteiches sequences with proteomes of fully sequenced eukaryotic pathogens, including fungi, apicomplexa and trypanosomatids, allowed the identification of A. euteiches genes with close orthologs in these microorganisms but absent in other oomycetes sequenced so far, notably transporters and non-ribosomal peptide synthetases, and suggests the presence of a defense mechanism against oxidative stress which was initially characterized in the pathogenic trypanosomatids.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
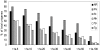
Figure 1. Comparison of A. euteiches sequences to the non redundant protein database at NCBI and proteomes of fully sequenced organisms.
Unigene sequences were blasted using the BLASTX software to the nr database and to the proteome of P. sojae (Ps), A. thaliana (At), N. haematococca (Nh), T. pseudonana (Tp) and T. gondii (Tg). For each class of e-value the percentage of unigenes showing homology is indicated.
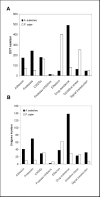
Figure 2. Classification of unigenes based on eight functional categories involved in pathogenicity.
A. euteiches unigenes (black bars) for which a putative function was assigned were classified into defined categories according to and . For each category the number of ESTs (A) and the number of unigene (B) is shown. For comparison to Phytophthora, analysis of a EST collection from P. sojae was added to the figure (white bars).
Figure 3. Protein domain organisation of cell surface proteins from oomycetes and apicomplexa.
Two proteins from Phytophthora, CBEL and CAR which are localized at the cell surface , are represented. CBEL contains two closely associated domains, a cellulose binding domain (CBD) and a N/apple PAN domain (PAN). CAR proteins are composed of a mucin-like region and a N-terminal region (CAR) with conserved cysteine residues. AMA1 is a P. falciparum protein with two PAN domains .
Figure 4. Growth of A. euteiches on synthetic media.
A. euteiches and Phytophthora parasitica mycelium were grown on synthetic medium containing Glucose (SM-Glc) or Pectin (SM-Pect) as carbon source. Photos were taken 15 days after inoculation.
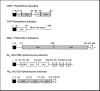
Figure 5. A cystatin-like protease inhibitor in A. euteiches.
A, domain organisation of the deduced protein sequence of Ae_2AL5945. A putative signal peptide (SP) and two domains similar to cystatin domain are shown. B, alignment of cystatin domains from P. infestans cystatins (EPIC1, EPIC2A, EPIC2B , EPIC3 and EPIC4) and from Ae_2AL5945. The active sites of cystatins, including the N-terminal trunk (NT), first binding loop (L1), and second binding loop (L2), are shown.
Figure 6. Identification of conserved amino acid residues at the N-terminal extremities of P. infestans and A. euteiches CRN sequences.
A, multiple alignment of A. euteiches like CRNs and P. infestans CRNs (CRN1, CRN5, CRN11, CRN14) showing the highest homology to A. euteiches sequences. B, consensus sequence pattern calculated using Weblogo showing the most conserved amino acid residues.
Figure 7. Identification of A. euteiches genes putatively involved in trypanothione synthesis.
A, trypanothione biosynthesis starts with the formation of glutathione catalysed by g-glutamyl synthetase and glutathione synthetase. Trypanothione is synthesized from glutathione and spermidine either in two successive steps catalysed by glutathionyl spermidine synthase and trypanothione synthase. It has been shown in some cases that trypanothione synthase is able to catalysed the two reactions. Unigenes predicted to code enzymes belonging to this pathway are indicated. B, multiple alignment of gluthationyl spermidine synthetase from E. coli (GSP_ECOLI), from Crythidia fasciculata (TRYS CRIFA) and Ae_2AL7562.
Similar articles
- Root rot disease of legumes caused by Aphanomyces euteiches.
Gaulin E, Jacquet C, Bottin A, Dumas B. Gaulin E, et al. Mol Plant Pathol. 2007 Sep;8(5):539-48. doi: 10.1111/j.1364-3703.2007.00413.x. Mol Plant Pathol. 2007. PMID: 20507520 - Sterol metabolism in the oomycete Aphanomyces euteiches, a legume root pathogen.
Madoui MA, Bertrand-Michel J, Gaulin E, Dumas B. Madoui MA, et al. New Phytol. 2009;183(2):291-300. doi: 10.1111/j.1469-8137.2009.02895.x. Epub 2009 Jun 3. New Phytol. 2009. PMID: 19496952 - Deciphering common and specific transcriptional immune responses in pea towards the oomycete pathogens Aphanomyces euteiches and Phytophthora pisi.
Hosseini S, Elfstrand M, Heyman F, Funck Jensen D, Karlsson M. Hosseini S, et al. BMC Genomics. 2015 Aug 21;16(1):627. doi: 10.1186/s12864-015-1829-1. BMC Genomics. 2015. PMID: 26293353 Free PMC article. - Emerging oomycete threats to plants and animals.
Derevnina L, Petre B, Kellner R, Dagdas YF, Sarowar MN, Giannakopoulou A, De la Concepcion JC, Chaparro-Garcia A, Pennington HG, van West P, Kamoun S. Derevnina L, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Dec 5;371(1709):20150459. doi: 10.1098/rstb.2015.0459. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 28080985 Free PMC article. Review. - A Meta-Analysis to Determine the State of Biological Control of Aphanomyces Root Rot.
Godebo AT, Wee NMJ, Yost CK, Walley FL, Germida JJ. Godebo AT, et al. Front Mol Biosci. 2022 Feb 2;8:777042. doi: 10.3389/fmolb.2021.777042. eCollection 2021. Front Mol Biosci. 2022. PMID: 35187066 Free PMC article. Review.
Cited by
- Arms race: diverse effector proteins with conserved motifs.
Liu L, Xu L, Jia Q, Pan R, Oelmüller R, Zhang W, Wu C. Liu L, et al. Plant Signal Behav. 2019;14(2):1557008. doi: 10.1080/15592324.2018.1557008. Epub 2019 Jan 9. Plant Signal Behav. 2019. PMID: 30621489 Free PMC article. - A molecular insight into algal-oomycete warfare: cDNA analysis of Ectocarpus siliculosus infected with the basal oomycete Eurychasma dicksonii.
Grenville-Briggs L, Gachon CM, Strittmatter M, Sterck L, Küpper FC, van West P. Grenville-Briggs L, et al. PLoS One. 2011;6(9):e24500. doi: 10.1371/journal.pone.0024500. Epub 2011 Sep 15. PLoS One. 2011. PMID: 21935414 Free PMC article. - The evolutionary phylogeny of the oomycete "fungi".
Beakes GW, Glockling SL, Sekimoto S. Beakes GW, et al. Protoplasma. 2012 Jan;249(1):3-19. doi: 10.1007/s00709-011-0269-2. Epub 2011 Mar 20. Protoplasma. 2012. PMID: 21424613 Review. - From pathogen genomes to host plant processes: the power of plant parasitic oomycetes.
Pais M, Win J, Yoshida K, Etherington GJ, Cano LM, Raffaele S, Banfield MJ, Jones A, Kamoun S, Saunders DG. Pais M, et al. Genome Biol. 2013 Jun 28;14(6):211. doi: 10.1186/gb-2013-14-6-211. Genome Biol. 2013. PMID: 23809564 Free PMC article. Review. - The elicitin-like glycoprotein, ELI025, is secreted by the pathogenic oomycete Pythium insidiosum and evades host antibody responses.
Lerksuthirat T, Lohnoo T, Inkomlue R, Rujirawat T, Yingyong W, Khositnithikul R, Phaonakrop N, Roytrakul S, Sullivan TD, Krajaejun T. Lerksuthirat T, et al. PLoS One. 2015 Mar 20;10(3):e0118547. doi: 10.1371/journal.pone.0118547. eCollection 2015. PLoS One. 2015. PMID: 25793767 Free PMC article.
References
- Delwich PA, Grau CR, Holub EB, Perry JB. Characterization of Aphanomyces euteiches isolates recovered from alfalfa in Wisconsin. Plant Dis. 1987;71:155–161.
- Levenfors JP, Wikstrom M, Persson L, Gerhardson B. Pathogenicity of Aphanomyces spp. from different leguminous crops in Sweden. Eur J Plant Pathol. 2003;109:535–543.
- Wicker E, Rouxel F. Specific behaviour of French Aphanomyces euteiches Drechs. populations for virulence and aggressiveness on pea, related to isolates from Europe, America and New Zealand. Eur J Plant Pathol. 2001;107:919–929.
- Mitchell JE, Yang CY. Factors affecting growth and development of Aphanomyces euteiches. . Phytopathology. 1966;56:917–922. - PubMed
- Larsson M. Pathogenicity, morphology and isozyme variability among isolates of Aphanomyces spp. from weeds and various plants. Mycol Res. 1994;98:231–240.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases