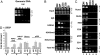Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development - PubMed (original) (raw)
Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development
Ming Yu et al. Proc Natl Acad Sci U S A. 2008.
Erratum in
- Proc Natl Acad Sci U S A. 2008 Apr 29;105(17):6502
Abstract
The CD4 gene is regulated in a stage-specific manner during T cell development, being repressed in CD4(-)CD8(-) double-negative (DN) and CD8 cells, but expressed in CD4(+)CD8(+) double-positive (DP) and CD4 cells. Furthermore, the expression/repression pattern is reversible in developing (DN and DP) thymocytes, but irreversible in mature (CD4 and CD8) T cells. Here, we explored the molecular mechanisms underlying this complex mode of regulation by examining the nucleoprotein structure of the CD4 locus throughout T cell development and in DN cells lacking the CD4 silencer. In DN cells, the CD4 enhancer is preloaded with multiple transcription activators, but p300 recruitment is impaired by the silencer that is associated with the repressor Runx1. DP cells achieve high-level CD4 expression via a combination of CD4 derepression and true activation, but Runx1 remains bound to the silencer that retains an open chromatin configuration. In CD4 cells, Runx1 dissociates from the silencer that has become less accessible, and CD4 transcription appears to be achieved via a mechanism distinct from that operating in DP cells. In CD8 cells, the CD4 promoter becomes incorporated into heterochromatin-like structure. Our data shed light on the molecular basis of CD4 regulation and provide a conceptual framework for understanding how the same regulatory elements can mediate both reversible and irreversible CD4 regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
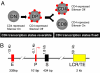
Regulation of CD4 expression during T cell development. (A) Expression of CD4 (small dots), in conjunction with that of CD8 (not shown), marks the three major stages in T cell development. CD4 is expressed in DP and CD4 cells, but repressed in DN and CD8 cells by the CD4 silencer. The repression/expression pattern in the developing thymocytes (DN and DP) is labile, but becomes irreversible once the cells mature into CD4/CD8 cells. (B) The regulatory elements in the CD4 locus. The CD4 promoter (P) and proximal enhancer (E) are each required to direct CD4 expression in all T cell subsets, whereas a thymic enhancer (TE) and an LCR are additionally required for high-level, uniform CD4 expression in DP cells in certain, but not all, transgenic models. The activities of the promoter/enhancers are suppressed by the CD4 silencer (S). The positions and sizes of the regulatory elements are indicated above and below the DNA, respectively.
Fig. 2.
ChIP analysis of the CD4 locus. (A) A multiplex PCR assay for semiquantitative measurement of the CD4 enhancer (E), silencer (S), promoter (P), and the GADPH gene (asterisk). The agarose gel is stained with SYBR green, which gives less background staining and a wider dynamic range than ethidium bromide. (B and C) ChIP assays showing histone modifications (B) and transcription factor binding (C) at the CD4 locus. Two million cell equivalents of chromatin were used for each immunoprecipitation, and 10% of precipitated DNA was analyzed by PCR. At least two independent sources of cells were used for chromatin preparation, and PCR was repeated at least three times for each precipitated DNA sample. Input, sonicated chromatin equivalent to 2,000 cells and, thus, representing 0.1% of the starting material; DN, double-negative thymocytes from Rag2−/− mice. (D) Quantitative analysis of ChIP samples. DNA from two experiments was quantified and the values averaged. Fold enrichment was then determined as described in Materials and Methods. R;S, DN3 cells isolated from Rag−/−; silencer−/− mice as described for Fig. 3.
Fig. 3.
Effects of CD4 silencer deletion in DN cells. (A) FACS analysis of thymocytes from mice with indicated genotypes. Total thymocytes (Total) are first resolved into DN3 and post-DN3 subsets based on CD25/CD44 expression. CD4/CD8 expression (columns 3 and 4) and cell sizes (column 5) of each subset are then compared. (B) The effects of CD4 silencer deletion on the CD4 locus in DN (Upper), CD8 (Lower), and DP (Lower) cells. Mi-2b, E2A, and HEB binding in Rag2−/−; silencer−/− cells was analyzed by multiplex PCR in parallel with the other lymphocyte subsets shown in Fig. 2_C_, and so the Rag2−/− control lanes were duplicated from the DN lanes in Fig. 2_C_.
Fig. 4.
The CD4 silencer is highly accessible in DP, but not CD4, cells. (A) Restriction enzyme map of the CD4 silencer and flanking regions. The position of the first nucleotide of the CD4 silencer sequence is set as +1, whereas the positions of the upstream sequences are given negative numbers. The enzymes for probing accessibility (BseR1, DdeI, Bsm I, AvaII, and PstI) are indicated above the DNA. Enzymes for generating normalization controls are indicated below the DNA, except that BseR1 (−327) and BseR I (784), which are used to probe the accessibility and thus indicated above the DNA, also are used to control for DdeI (386) and DdeI (782), respectively. Note that the BseR I cleavage site is 10 base pairs away from the recognition site, allowing BseR I (784) to control for DdeI (782) (Right Middle). The arrowheads denote the nested primers used for primer extension in the last step of LM-PCR. The illustration was generated by using the GCK2 software. (B) Accessibility of the CD4 silencer and flanking regions to BseR1, DdeI, and Bsm I. The arrowheads denote cleavage products, with the numbers referring to the positions of the enzyme recognition sites. (C) The intensities of the amplicons in Fig. 4_B_ were quantified, and the accessibility, defined as the intensities of the short relative to the long amplicons, was calculated. AvaII and PstI digestions were similarly quantified (data not shown). The accessibility in CD4 relative to DP cells was then plotted against the positions of the restriction enzyme sites. The numbers on the x axis are as defined in A. All of the sites depicted in A are included except for BseR (−327), which is resistant to digestion (data not shown). The accessibility results for the CD4 silencer and its flanking sequences were reproduced in at least two independent experiments (see also
SI Fig. 6_A_
).
Fig. 5.
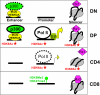
A model for nucleoprotein organization at the CD4 enhancer, promoter, and silencer in various T cell subsets. For simplicity, the CD4 enhancer and promoter are depicted as naked DNA instead of nucleosomes, and Mi-2b and p300 are shown to bind only the enhancer, although they also bind the promoter and silencer in DP cells. The dotted line circling p300 in DN cells signifies low occupancy, as is the dotted outline of Pol II in CD4 cells. Mi-2b, HEB, and p300 binding as well as histone acetylation at the endogenous CD4 locus in DP cells are consistent with the data from a previous study using total thymocytes (11), whereas the binding of Runx3 in CD8 cells is based solely on a previous article (23).
Similar articles
- Thymocytes control the CD4 gene differently from mature T lymphocytes.
Uematsu Y, Donda A, De Libero G. Uematsu Y, et al. Int Immunol. 1997 Jan;9(1):179-87. doi: 10.1093/intimm/9.1.179. Int Immunol. 1997. PMID: 9043959 - A general approach for controlling transcription and probing epigenetic mechanisms: application to the CD4 locus.
Wan M, Kaundal R, Huang H, Zhao J, Yang X, Chaiyachati BH, Li S, Chi T. Wan M, et al. J Immunol. 2013 Jan 15;190(2):737-47. doi: 10.4049/jimmunol.1201278. J Immunol. 2013. PMID: 23293358 Free PMC article. - Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex.
Wan M, Zhang J, Lai D, Jani A, Prestone-Hurlburt P, Zhao L, Ramachandran A, Schnitzler GR, Chi T. Wan M, et al. Eur J Immunol. 2009 Feb;39(2):580-8. doi: 10.1002/eji.200838909. Eur J Immunol. 2009. PMID: 19180471 Free PMC article. - The regulation of CD4 and CD8 coreceptor gene expression during T cell development.
Ellmeier W, Sawada S, Littman DR. Ellmeier W, et al. Annu Rev Immunol. 1999;17:523-54. doi: 10.1146/annurev.immunol.17.1.523. Annu Rev Immunol. 1999. PMID: 10358767 Review. - Controlling CD4 gene expression during T cell lineage commitment.
Siu G. Siu G. Semin Immunol. 2002 Dec;14(6):441-51. doi: 10.1016/s1044532302000799. Semin Immunol. 2002. PMID: 12457617 Review.
Cited by
- Enhancers in T Cell development and malignant lesions.
Zhang T, Zou L. Zhang T, et al. Cell Death Discov. 2024 Sep 17;10(1):406. doi: 10.1038/s41420-024-02160-7. Cell Death Discov. 2024. PMID: 39284807 Free PMC article. Review. - The histone chaperone CAF-1 cooperates with the DNA methyltransferases to maintain Cd4 silencing in cytotoxic T cells.
Ng C, Aichinger M, Nguyen T, Au C, Najar T, Wu L, Mesa KR, Liao W, Quivy JP, Hubert B, Almouzni G, Zuber J, Littman DR. Ng C, et al. Genes Dev. 2019 Jun 1;33(11-12):669-683. doi: 10.1101/gad.322024.118. Epub 2019 Apr 11. Genes Dev. 2019. PMID: 30975723 Free PMC article. - Heritable Gene Regulation in the CD4:CD8 T Cell Lineage Choice.
Issuree PD, Ng CP, Littman DR. Issuree PD, et al. Front Immunol. 2017 Mar 22;8:291. doi: 10.3389/fimmu.2017.00291. eCollection 2017. Front Immunol. 2017. PMID: 28382035 Free PMC article. Review. - Distinct requirement of Runx complexes for TCRβ enhancer activation at distinct developmental stages.
Seo W, Muroi S, Akiyama K, Taniuchi I. Seo W, et al. Sci Rep. 2017 Feb 2;7:41351. doi: 10.1038/srep41351. Sci Rep. 2017. PMID: 28150718 Free PMC article. - LMO2 induces T-cell leukemia with epigenetic deregulation of CD4.
Cleveland SM, Goodings C, Tripathi RM, Elliott N, Thompson MA, Guo Y, Shyr Y, Davé UP. Cleveland SM, et al. Exp Hematol. 2014 Jul;42(7):581-93.e5. doi: 10.1016/j.exphem.2014.04.010. Epub 2014 May 2. Exp Hematol. 2014. PMID: 24792354 Free PMC article.
References
- Groettrup M, von Boehmer H. A role for a pre-T-cell receptor in T-cell development. Immunol Today. 1993;14:610–614. - PubMed
- Bosselut R. CD4/CD8-lineage differentiation in the thymus: From nuclear effectors to membrane signals. Nat Rev Immunol. 2004;4:529–540. - PubMed
- Singer A, Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: Analysis of the CD4/CD8 lineage decision. Adv Immunol. 2004;83:91–131. - PubMed
- Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: New insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. - PubMed
- Zou YR, et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous