Consistent blind protein structure generation from NMR chemical shift data - PubMed (original) (raw)
. 2008 Mar 25;105(12):4685-90.
doi: 10.1073/pnas.0800256105. Epub 2008 Mar 7.
Oliver Lange, Frank Delaglio, Paolo Rossi, James M Aramini, Gaohua Liu, Alexander Eletsky, Yibing Wu, Kiran K Singarapu, Alexander Lemak, Alexandr Ignatchenko, Cheryl H Arrowsmith, Thomas Szyperski, Gaetano T Montelione, David Baker, Ad Bax
Affiliations
- PMID: 18326625
- PMCID: PMC2290745
- DOI: 10.1073/pnas.0800256105
Consistent blind protein structure generation from NMR chemical shift data
Yang Shen et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Protein NMR chemical shifts are highly sensitive to local structure. A robust protocol is described that exploits this relation for de novo protein structure generation, using as input experimental parameters the (13)C(alpha), (13)C(beta), (13)C', (15)N, (1)H(alpha) and (1)H(N) NMR chemical shifts. These shifts are generally available at the early stage of the traditional NMR structure determination process, before the collection and analysis of structural restraints. The chemical shift based structure determination protocol uses an empirically optimized procedure to select protein fragments from the Protein Data Bank, in conjunction with the standard ROSETTA Monte Carlo assembly and relaxation methods. Evaluation of 16 proteins, varying in size from 56 to 129 residues, yielded full-atom models that have 0.7-1.8 A root mean square deviations for the backbone atoms relative to the experimentally determined x-ray or NMR structures. The strategy also has been successfully applied in a blind manner to nine protein targets with molecular masses up to 15.4 kDa, whose conventional NMR structure determination was conducted in parallel by the Northeast Structural Genomics Consortium. This protocol potentially provides a new direction for high-throughput NMR structure determination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
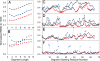
Plots of normalized accuracy of database fragments selected for ubiquitin. For each ubiquitin segment, 200 fragment candidates of the same length were selected using either the standard ROSETTA procedure (filled triangles), or an MFR search of the 5665-protein structural database, assigned by the programs DC (filled circles) or SPARTA (filled diamonds). For all panels, coordinate rmsds (N, Cα, and C′) between query segment and selected fragments are normalized with respect to randomly selected fragments. (A and B) Average (A) and lowest (B) normalized rmsd of 200 selected fragments, as a function of fragment size, relative to the x-ray coordinates of the corresponding ubiquitin segment, averaged over all (overlapped) consecutive segments. (C and D) Average normalized rmsd of 200 nine-residue (C) and three-residue (D) fragments relative to the x-ray coordinates, as a function of position in the ubiquitin sequence. (E and F) Lowest normalized rmsd of any of these selected nine-residue (E) or three-residue (F) fragments.
Fig. 2.
Plots of ROSETTA all atom energy versus Cα rmsd relative to the experimental structures for four representative test proteins. (A–D) Standard ROSETTA all atom energy. (A′–D′) ROSETTA energy, rescored by using the experimental chemical shifts (Eq. 1). (A) Ubiquitin. (B) Calbindin. (C) HPr. (D) TM1112. For A′–D′, the model with the lowest energy, marked by an arrow, is shown in Fig. 3 or
SI Fig. 9
.
Fig. 3.
Backbone ribbon representations (32) of the lowest-energy CS-ROSETTA structure (red) superimposed on the experimental x-ray/NMR structures (blue), with superposition optimized for ordered residues, as defined in the footnote to
SI Table 3
. (A) GB3. (B) CspA. (C) Calbindin. (D) Ubiquitin. (E) DinI. (F) Apo_lafbp. Overlays of the 10 remaining structures are shown in
SI Fig. 9
.
Fig. 4.
Results from blind CS-ROSETTA structure generation for four structural genomics targets (Table 2). The remaining five are in
SI Fig. 12
. (A–D) Superposition of lowest-energy CS-ROSETTA models (red) with experimental NMR structures (blue), with superposition optimized for ordered residues, as defined in the footnote to
SI Table 5
. (E–H) Plots of rescored (Eq. 1) ROSETTA all-atom energy versus Cα rmsd relative to the lowest-energy model (bold dot on vertical axis). (A and E) StR82. (B and F) RpT7. (C and G) VfR117. (D and H) NeT4.
Comment in
- Local knowledge helps determine protein structures.
Gryk MR, Hoch JC. Gryk MR, et al. Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4533-4. doi: 10.1073/pnas.0801069105. Epub 2008 Mar 19. Proc Natl Acad Sci U S A. 2008. PMID: 18353980 Free PMC article. No abstract available. - Structural biology. Protein structure gets exciting.
Doerr A. Doerr A. Nat Methods. 2010 Nov;7(11):870-1. doi: 10.1038/nmeth1110-870a. Nat Methods. 2010. PMID: 21049576
Similar articles
- De novo protein structure generation from incomplete chemical shift assignments.
Shen Y, Vernon R, Baker D, Bax A. Shen Y, et al. J Biomol NMR. 2009 Feb;43(2):63-78. doi: 10.1007/s10858-008-9288-5. Epub 2008 Nov 26. J Biomol NMR. 2009. PMID: 19034676 Free PMC article. - CABS-NMR--De novo tool for rapid global fold determination from chemical shifts, residual dipolar couplings and sparse methyl-methyl NOEs.
Latek D, Kolinski A. Latek D, et al. J Comput Chem. 2011 Feb;32(3):536-44. doi: 10.1002/jcc.21640. Epub 2010 Aug 30. J Comput Chem. 2011. PMID: 20806263 - Rapid protein fold determination using unassigned NMR data.
Meiler J, Baker D. Meiler J, et al. Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15404-9. doi: 10.1073/pnas.2434121100. Epub 2003 Dec 10. Proc Natl Acad Sci U S A. 2003. PMID: 14668443 Free PMC article. - Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures.
Mao B, Tejero R, Baker D, Montelione GT. Mao B, et al. J Am Chem Soc. 2014 Feb 5;136(5):1893-906. doi: 10.1021/ja409845w. Epub 2014 Jan 23. J Am Chem Soc. 2014. PMID: 24392845 Free PMC article. - Chemical shift-based methods in NMR structure determination.
Nerli S, McShan AC, Sgourakis NG. Nerli S, et al. Prog Nucl Magn Reson Spectrosc. 2018 Jun-Aug;106-107:1-25. doi: 10.1016/j.pnmrs.2018.03.002. Epub 2018 Mar 11. Prog Nucl Magn Reson Spectrosc. 2018. PMID: 31047599 Free PMC article. Review.
Cited by
- Molecular Characterisation of Titin N2A and Its Binding of CARP Reveals a Titin/Actin Cross-linking Mechanism.
Zhou T, Fleming JR, Lange S, Hessel AL, Bogomolovas J, Stronczek C, Grundei D, Ghassemian M, Biju A, Börgeson E, Bullard B, Linke WA, Chen J, Kovermann M, Mayans O. Zhou T, et al. J Mol Biol. 2021 Apr 30;433(9):166901. doi: 10.1016/j.jmb.2021.166901. Epub 2021 Feb 27. J Mol Biol. 2021. PMID: 33647290 Free PMC article. - Protein structure determination by combining sparse NMR data with evolutionary couplings.
Tang Y, Huang YJ, Hopf TA, Sander C, Marks DS, Montelione GT. Tang Y, et al. Nat Methods. 2015 Aug;12(8):751-4. doi: 10.1038/nmeth.3455. Epub 2015 Jun 29. Nat Methods. 2015. PMID: 26121406 Free PMC article. - NMR determines transient structure and dynamics in the disordered C-terminal domain of WASp interacting protein.
Haba NY, Gross R, Novacek J, Shaked H, Zidek L, Barda-Saad M, Chill JH. Haba NY, et al. Biophys J. 2013 Jul 16;105(2):481-93. doi: 10.1016/j.bpj.2013.05.046. Biophys J. 2013. PMID: 23870269 Free PMC article. - Prediction of Protein Structure Using Surface Accessibility Data.
Hartlmüller C, Göbl C, Madl T. Hartlmüller C, et al. Angew Chem Int Ed Engl. 2016 Sep 19;55(39):11970-4. doi: 10.1002/anie.201604788. Epub 2016 Aug 25. Angew Chem Int Ed Engl. 2016. PMID: 27560616 Free PMC article. - Segmental isotope labeling of proteins for NMR structural study using a protein S tag for higher expression and solubility.
Kobayashi H, Swapna GV, Wu KP, Afinogenova Y, Conover K, Mao B, Montelione GT, Inouye M. Kobayashi H, et al. J Biomol NMR. 2012 Apr;52(4):303-13. doi: 10.1007/s10858-012-9610-0. Epub 2012 Mar 3. J Biomol NMR. 2012. PMID: 22389115 Free PMC article.
References
- Atreya HS, Szyperski T. Rapid NMR data collection. Methods Enzymol. 2005;394:78–108. - PubMed
- Freeman R, Kupce E. New methods for fast multidimensional NMR. J Biomol NMR. 2003;27:101–113. - PubMed
- Wagner G, Pardi A, Wüthrich K. Hydrogen-bond length and H-1-NMR chemical-shifts in proteins. J Am Chem Soc. 1983;105:5948–5949.
- Williamson MP, Asakura T. Empirical comparisons of models for chemical-shift calculation in proteins. J Magn Reson B. 1993;101:63–71.
- Case DA. Calibration of ring-current effects in proteins and nucleic acids. J Biomol NMR. 1995;6:341–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM074958/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- ImNIH/Intramural NIH HHS/United States
- U54-GM074958/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources