Phosphoproteome analysis of Drosophila melanogaster embryos - PubMed (original) (raw)
Phosphoproteome analysis of Drosophila melanogaster embryos
Bo Zhai et al. J Proteome Res. 2008 Apr.
Abstract
Protein phosphorylation is a key regulatory event in most cellular processes and development. Mass spectrometry-based proteomics provides a framework for the large-scale identification and characterization of phosphorylation sites. Here, we used a well-established phosphopeptide enrichment and identification strategy including the combination of strong cation exchange chromatography, immobilized metal affinity chromatography, and high-accuracy mass spectrometry instrumentation to study phosphorylation in developing Drosophila embryos. In total, 13,720 different phosphorylation sites were discovered from 2702 proteins with an estimated false-discovery rate (FDR) of 0.63% at the peptide level. Because of the large size of the data set, both novel and known phosphorylation motifs were extracted using the Motif-X algorithm, including those representative of potential ordered phosphorylation events.
Figures
Figure 1
Schematic illustration of the strategy for large-scale phosphorylation site identification from Drosophila embryos. (A) The 0–24 h old D. melanogaster w118 embryos were lysed and directly digested with trypsin. Tryptic peptides were desalted and then separated by SCX chromatography. Phosphopeptides from 12 SCX fractions were further enriched by IMAC and then analyzed by LC–MS/MS techniques. (B) MS/MS spectra from 24 analyses (duplicates for each sample) were searched against a composite target-decoy Drosophila protein database. Mass deviation, XCorr, dCn′, and solution charge state were used to filter correct from incorrect matches, maintaining <1% false-discovery rate (FDR). In total, 36 203 phosphopeptides (16 822 unique phosphopeptides) and 13 720 nonredundant phosphorylation sites were identified at a FDR of 0.63% (229 decoy matches). High-certainty localization (Ascore ≥ 13; P ≤0.05) was found for 10 038 sites. Finally, phosphorylation motifs (standard, degenerate, and multiply phosphorylated) were extracted from the data set with the Motif-X algorithm.
Figure 2
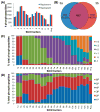
Distributions of phosphopeptides and their properties across 12 SCX fractions. (A) The number of phosphopeptides identified from duplicate analyses of each fraction. While similar numbers of peptides were identified in each replicate, an average of 41.3 ± 6.8% more peptides could be attributed solely to analyzing each fraction twice. (B) Venn diagram depicting the extent of overlap for the phosphopeptides identified in duplicate analyses of fraction 3. Numbers in parentheses indicate the percentage of either replicate that lies outside the overlap region. (C) Nonredundant phosphopeptides in each fraction (and replicate) with calculated solution charge states between −1 and +4. SCX separates phosphopeptides based primarily on solution charge. (D) Nonredundant phosphopeptides in each fraction containing 1 (1P) to 6 (6P) phosphorylation sites. (a and b correspond to duplicate LC–MS/MS runs of each SCX fraction).
Figure 3
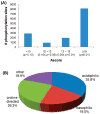
(A) Ascore distribution for all identified sites from 36 203 peptides. Most sites could be localized with near (P ≤ 0.01) or high (P ≤ 0.05) certainty. (B) Classification of phosphorylation events into 4 general sequence categories based on kinase specificities.
Figure 4
Logo-like representations of acidic phosphorylation motifs identified by the Motif-X algorithm. (A) Examples of motifs extracted utilizing all 20 amino acids. Note that several permutations of a similar motif are identified. (B) Degenerate motif for casein-kinase II-like phosphorylation. Note that all three motifs in panel A are represented in this single motif. The residue at position +4 is also now significant. (C) Acidic, double phosphorylation motif identified using both degenerate analysis and considering multiple phosphorylation events. Phosphorylated serine and threonine (denoted as B and X, respectively) function as an acidic residue at position +1 in this motif.
Figure 5
Examples of motifs (represented by logo-like representations) identified in this study. The phosphorylated serine, threonine, or tyrosine are centered. (A) PKA-like substrate motifs showing a preference for arginine over lysine in degenerate analyses. (B) Novel threonine single phosphorylation motif. (C) Double phosphorylation motif (B and X represent phosphoserine and threonine, respectively) suggestive of ordered phosphorylation. (D) Examples of tyrosine single phosphorylation motifs.
Figure 6
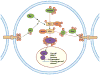
Overlay example of phosphorylation sites from this study on a developmental pathway. The Salvador–Warts–Hippo (SWH) pathway controls organ size by modulating cell growth, proliferation, and apoptosis. Many of the genetic and biochemical interactions are known, but post-translational modifications such as phosphorylation affecting core components are almost entirely lacking. Phosphorylation events detected in this study are overlaid onto a representation of the pathway. Yorkie (Yki) is hyperphosphorylated in Drosophila embryos. The arrows in the figure represent what is known about the pathway from the literature where a given protein acts to stimulate or inhibit the function of the pathway. They do not indicate the direct kinase–substrate relationship.
Similar articles
- Improvement of phosphoproteome analyses using FAIMS and decision tree fragmentation. application to the insulin signaling pathway in Drosophila melanogaster S2 cells.
Bridon G, Bonneil E, Muratore-Schroeder T, Caron-Lizotte O, Thibault P. Bridon G, et al. J Proteome Res. 2012 Feb 3;11(2):927-40. doi: 10.1021/pr200722s. Epub 2011 Dec 1. J Proteome Res. 2012. PMID: 22059388 - Online automated in vivo zebrafish phosphoproteomics: from large-scale analysis down to a single embryo.
Lemeer S, Pinkse MW, Mohammed S, van Breukelen B, den Hertog J, Slijper M, Heck AJ. Lemeer S, et al. J Proteome Res. 2008 Apr;7(4):1555-64. doi: 10.1021/pr700667w. Epub 2008 Feb 29. J Proteome Res. 2008. PMID: 18307296 - Quantitative phosphoproteome analysis of Streptomyces coelicolor by immobilized zirconium (IV) affinity chromatography and mass spectrometry reveals novel regulated protein phosphorylation sites and sequence motifs.
Alonso-Fernández S, Arribas-Díez I, Fernández-García G, González-Quiñónez N, Jensen ON, Manteca A. Alonso-Fernández S, et al. J Proteomics. 2022 Oct 30;269:104719. doi: 10.1016/j.jprot.2022.104719. Epub 2022 Sep 8. J Proteomics. 2022. PMID: 36089190 - Enrichment Strategies in Phosphoproteomics.
Leitner A. Leitner A. Methods Mol Biol. 2016;1355:105-21. doi: 10.1007/978-1-4939-3049-4_7. Methods Mol Biol. 2016. PMID: 26584921 Review. - Tools for analyzing the phosphoproteome and other phosphorylated biomolecules: a review.
Leitner A, Sturm M, Lindner W. Leitner A, et al. Anal Chim Acta. 2011 Oct 3;703(1):19-30. doi: 10.1016/j.aca.2011.07.012. Epub 2011 Jul 19. Anal Chim Acta. 2011. PMID: 21843671 Review.
Cited by
- PredPhos: an ensemble framework for structure-based prediction of phosphorylation sites.
Gao Y, Hao W, Gu J, Liu D, Fan C, Chen Z, Deng L. Gao Y, et al. J Biol Res (Thessalon). 2016 Jul 4;23(Suppl 1):12. doi: 10.1186/s40709-016-0042-y. eCollection 2016 May. J Biol Res (Thessalon). 2016. PMID: 27437197 Free PMC article. - Differential phosphoproteomics of fibroblast growth factor signaling: identification of Src family kinase-mediated phosphorylation events.
Cunningham DL, Sweet SM, Cooper HJ, Heath JK. Cunningham DL, et al. J Proteome Res. 2010 May 7;9(5):2317-28. doi: 10.1021/pr9010475. J Proteome Res. 2010. PMID: 20225815 Free PMC article. - Cyclin Y is a novel conserved cyclin essential for development in Drosophila.
Liu D, Finley RL Jr. Liu D, et al. Genetics. 2010 Apr;184(4):1025-35. doi: 10.1534/genetics.110.114017. Epub 2010 Jan 25. Genetics. 2010. PMID: 20100936 Free PMC article. - Discovery of protein phosphorylation motifs through exploratory data analysis.
Chen YC, Aguan K, Yang CW, Wang YT, Pal NR, Chung IF. Chen YC, et al. PLoS One. 2011;6(5):e20025. doi: 10.1371/journal.pone.0020025. Epub 2011 May 25. PLoS One. 2011. PMID: 21647451 Free PMC article. - Drosophila mbm is a nucleolar myc and casein kinase 2 target required for ribosome biogenesis and cell growth of central brain neuroblasts.
Hovhanyan A, Herter EK, Pfannstiel J, Gallant P, Raabe T. Hovhanyan A, et al. Mol Cell Biol. 2014 May;34(10):1878-91. doi: 10.1128/MCB.00658-13. Epub 2014 Mar 10. Mol Cell Biol. 2014. PMID: 24615015 Free PMC article.
References
- Celniker SE, Rubin GM. The Drosophila melanogaster genome. Ann Rev Genomics Hum Genet. 2003;4(1):89–117. - PubMed
- Adams MD. The genome sequence of Drosophila melanogaster. Science. 2000;218:5–2195. - PubMed
- Peter Bangs KW. Regulation and execution of apoptosis during Drosophila development. Dev Dyn. 2000;218(1):68–79. - PubMed
- Vidal M, Cagan RL. Drosophila models for cancer research. Curr Opin Genet Dev. 2006;16(1):10–16. - PubMed
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Ann Rev Cell Dev Biol. 2006;22(1):623–650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases