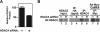Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4 - PubMed (original) (raw)
Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4
Johannes Backs et al. Mol Cell Biol. 2008 May.
Abstract
Calcium/calmodulin-dependent protein kinase II (CaMKII) phosphorylates histone deacetylase 4 (HDAC4), a class IIa HDAC, resulting in the cytosolic accumulation of HDAC4 and the derepression of the transcription factor myocyte enhancer factor 2. Phosphorylation by CaMKII requires docking of the kinase to a specific domain of HDAC4 not present in other HDACs. Paradoxically, however, CaMKII signaling can also promote the nuclear export of other class IIa HDACs, such as HDAC5. Here, we show that HDAC4 and HDAC5 form homo- and hetero-oligomers via a conserved coiled-coil domain near their amino termini. Whereas HDAC5 alone is unresponsive to CaMKII, it becomes responsive to CaMKII in the presence of HDAC4. The acquisition of CaMKII responsiveness by HDAC5 is mediated by HDAC5's direct association with HDAC4 and can occur by phosphorylation of HDAC4 or by transphosphorylation by CaMKII bound to HDAC4. Thus, HDAC4 integrates upstream Ca(2+)-dependent signals via its association with CaMKII and transmits these signals to HDAC5 by protein-protein interactions. We conclude that HDAC4 represents a point of convergence for CaMKII signaling to downstream HDAC-regulated genes, and we suggest that modulation of the interaction of CaMKII and HDAC4 represents a means of regulating CaMKII-dependent gene programs.
Figures
FIG. 1.
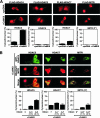
Coshuttling of HDAC4 and HDAC5. (A) COS cells were transfected with expression plasmids encoding FLAG-HDAC4, -HDAC5, -HDAC7, and -MITR (referred to as 9*) in the absence or presence of expression plasmids encoding constitutively active CaMKII, or they were transfected with pCDNA1 as a control. The subcellular localization of these HDACs was determined 1 day after transfection by immunostaining for the FLAG epitope tag. CaMKII induced complete cytosolic accumulation of HDAC4 and partial nuclear export of HDAC7. (B) Green fluorescent protein (GFP)-HDAC4 was coexpressed with FLAG-HDAC5, -HDAC7, and -MITR in COS cells. HDAC4 colocalized with HDAC5 and MITR. CaMKII induced the nuclear export of HDAC5 and, to a lesser degree, of MITR in the presence of HDAC4 but not in its absence. The percentage of cells showing the localization of the indicated HDAC in the cytoplasm relative to that in the nucleus when cotransfected with pcDNA or activated CaMKII is shown beneath each set of panels. At least 100 HDAC-expressing cells were analyzed under each condition. The data are expressed as the means ± SEMs of the results for three randomly chosen areas per slide. In panel B, HDAC4 was coexpressed with HDAC5, HDAC7, or MITR at 1:1 and 3:1 ratios, as indicated.
FIG. 2.
HDAC4 hetero-oligomerizes with HDAC5. (A) COS cells were transfected with expression plasmids encoding epitope-tagged HDACs, as indicated. Immunoprecipitation (IP) with anti-FLAG followed by immunoblotting (IB) with anti-Myc revealed interactions between HDAC4, HDAC5, and MITR (referred to as 9*). The middle and lower panels show input proteins as detected by IB. (B) COS cells were transfected with expression plasmids encoding Myc-HDAC5 and FLAG-HDAC4 and various deletion mutants of FLAG-HDAC4. IP followed by IB revealed that HDAC5 binds to HDAC4 through its N-terminal extension. (C) Sequence alignment of the N-terminal regions of HDAC4, -5, -7, and -9. (D) COS cells were transfected with expression plasmids encoding Myc-MEF2C, -CtBP, -HDAC4, -HDAC5, FLAG-HDAC4 wild type (WT), and FLAG-HDAC4 mutants lacking the coiled-coil domain (Δcc) and the MEF2 binding domain (ΔMEF2). Protein-protein interactions were detected by IP followed by IB. (E) Schematic diagram of HDAC4 and deletion mutants and summary of interactions with HDAC5 as detected by coimmunoprecipitation. NLS, nuclear localization sequence; NES, nuclear export sequence.
FIG. 3.
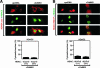
Hetero-oligomerization and CaMKII docking are required for coshuttling. FLAG-HDAC4 lacking the coiled-coil domain (Δcc) (A) or encoding a disrupted CaMKII docking site (R601F) (B) was expressed in COS cells together with GFP-HDAC5. The coiled-coil domain deletion abolished the colocalization of HDAC4 with HDAC5 and prevented the coshuttling of HDAC5 to the cytosol in response to constitutively active CaMKII. Mutating the CaMKII docking site did not affect colocalization with HDAC5 in the nucleus but did prevent CaMKII responsiveness of the HDAC4-HDAC5 complex. The percentage of cells showing HDAC5 in the cytoplasm when cotransfected with HDAC4 Δcc or HDAC4 R601F and activated CaMKII is shown beneath each set of panels. At least 100 HDAC-expressing cells were analyzed under each condition. The data are expressed as the means ± SEMs of the results for three randomly chosen areas per slide. The HDAC4 mutants were coexpressed with HDAC5 at 1:1 and 3:1 ratios, as indicated.
FIG. 4.
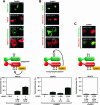
“Piggyback” phosphorylation versus transphosphorylation by CaMKII. (A) Green fluorescent protein (GFP)-HDAC5 lacking the phosphorylation sites Ser-259 and -498 (HDAC5-S/A; green) and FLAG-HDAC4 (red) were expressed separately (a and b) or together (c and d) in COS cells with constitutively active CaMKII. HDAC5-S/A did not respond to CaMKII alone but was exported from the nucleus to the cytoplasm when expressed together with FLAG-HDAC4. (B) GFP-HDAC5 (green) and FLAG-HDAC4-S/A (in which serines 246, 467, and 632 were replaced with alanine) (red) were expressed separately (a and b) or together (c and d) in COS cells with constitutively active CaMKII. Neither of the HDACs alone can respond to CaMKII, but when expressed together, both were exported from the nucleus to the cytoplasm in response to CaMKII. The percentage of cells showing HDAC5 in the cytoplasm when cotransfected with HDAC4 and activated CaMKII is shown beneath each set of panels. At least 100 HDAC-expressing cells were analyzed under each condition. The data are expressed as the means ± SEMs of the results for three randomly chosen areas per slide. HDAC4 was coexpressed with HDAC5 at 1:1, 3:1, and 1:3 ratios, as indicated. (C) When GFP-HDAC5-S/A and FLAG-HDAC4-S/A were coexpressed in COS cells, neither protein was exported from the nucleus to the cytoplasm in response to CaMKII. Schematic diagrams of the interactions of HDAC4 and HDAC5 are shown in each panel.
FIG. 5.
Interaction of endogenous HDAC4 and HDAC5 in C2C12 myoblasts. (A) The exposure of C2C12 myoblasts to HDAC4 siRNA achieved ∼50% downregulation of endogenous HDAC4 protein, as determined by immunoblot (IB) analysis and quantification by ImageJ software. (B) C2C12 myoblasts were treated with HDAC4 siRNA, as indicated. “Input” shows the level of endogenous HDAC5 (lanes 1 and 2). IP:HA designates a control IP using antihemagglutinin antibody to show background (bgrd) (lanes 3 and 4). Endogenous HDAC4 was immunoprecipitated (IP), and bound HDAC5 was determined by IB under control conditions and after HDAC4 knockdown (lanes 5 and 6). C2C12 cells were also infected with adenovirus (Ad) encoding Myc-CaMKII. Following IP with anti-Myc, the presence of HDAC5 in the CaMKII complex was determined by IB for endogenous HDAC5 (lanes 7 and 8).
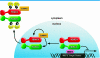
FIG. 6.
Model of CaMKII- and HDAC4-dependent nuclear export of HDAC5. CaMKII interacts with a specific docking site on HDAC4, which results in the phosphorylation of HDAC4 and 14-3-3 protein-mediated nuclear export in response to signals that activate CaMKII. While CaMKII does not bind directly to HDAC5, hetero-oligomerization of HDAC5 with HDAC4 enables CaMKII to transphosphorylate HDAC5. 14-3-3 protein binding to either HDAC4 or HDAC5 is sufficient to induce nuclear export of the HDAC4-HDAC5 complex with consequent activation of transcription factors, such as MEF2, which are repressed by class IIa HDACs.
Similar articles
- Nuclear calcium/calmodulin-dependent protein kinase IIdelta preferentially transmits signals to histone deacetylase 4 in cardiac cells.
Little GH, Bai Y, Williams T, Poizat C. Little GH, et al. J Biol Chem. 2007 Mar 9;282(10):7219-31. doi: 10.1074/jbc.M604281200. Epub 2006 Dec 19. J Biol Chem. 2007. PMID: 17179159 - CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy.
Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. Backs J, et al. J Clin Invest. 2006 Jul;116(7):1853-64. doi: 10.1172/JCI27438. Epub 2006 Jun 8. J Clin Invest. 2006. PMID: 16767219 Free PMC article. - Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5.
McKinsey TA, Zhang CL, Olson EN. McKinsey TA, et al. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14400-5. doi: 10.1073/pnas.260501497. Proc Natl Acad Sci U S A. 2000. PMID: 11114197 Free PMC article. - Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review. - Roles of Histone Deacetylase 4 in the Inflammatory and Metabolic Processes.
Kang H, Park YK, Lee JY, Bae M. Kang H, et al. Diabetes Metab J. 2024 May;48(3):340-353. doi: 10.4093/dmj.2023.0174. Epub 2024 Mar 22. Diabetes Metab J. 2024. PMID: 38514922 Free PMC article. Review.
Cited by
- Aurora B-dependent regulation of class IIa histone deacetylases by mitotic nuclear localization signal phosphorylation.
Guise AJ, Greco TM, Zhang IY, Yu F, Cristea IM. Guise AJ, et al. Mol Cell Proteomics. 2012 Nov;11(11):1220-9. doi: 10.1074/mcp.M112.021030. Epub 2012 Aug 2. Mol Cell Proteomics. 2012. PMID: 22865920 Free PMC article. - Endurance Training Regulates Expression of Some Angiogenesis-Related Genes in Cardiac Tissue of Experimentally Induced Diabetic Rats.
Khajehlandi M, Bolboli L, Siahkuhian M, Rami M, Tabandeh M, Khoramipour K, Suzuki K. Khajehlandi M, et al. Biomolecules. 2021 Mar 25;11(4):498. doi: 10.3390/biom11040498. Biomolecules. 2021. PMID: 33806202 Free PMC article. - Class IIa HDACs inhibit cell death pathways and protect muscle integrity in response to lipotoxicity.
Martin SD, Connor T, Sanigorski A, McEwen KA, Henstridge DC, Nijagal B, De Souza D, Tull DL, Meikle PJ, Kowalski GM, Bruce CR, Gregorevic P, Febbraio MA, Collier FM, Walder KR, McGee SL. Martin SD, et al. Cell Death Dis. 2023 Dec 1;14(12):787. doi: 10.1038/s41419-023-06319-5. Cell Death Dis. 2023. PMID: 38040704 Free PMC article. - Emerging roles for histone deacetylases in pulmonary hypertension and right ventricular remodeling (2013 Grover Conference series).
Cavasin MA, Stenmark KR, McKinsey TA. Cavasin MA, et al. Pulm Circ. 2015 Mar;5(1):63-72. doi: 10.1086/679700. Pulm Circ. 2015. PMID: 25992271 Free PMC article. Review. - Exercise-induced histone modifications in human skeletal muscle.
McGee SL, Fairlie E, Garnham AP, Hargreaves M. McGee SL, et al. J Physiol. 2009 Dec 15;587(Pt 24):5951-8. doi: 10.1113/jphysiol.2009.181065. J Physiol. 2009. PMID: 19884317 Free PMC article.
References
- Backs, J., and E. N. Olson. 2006. Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 9815-24. - PubMed
- Berdeaux, R., N. Goebel, L. Banaszynski, H. Takemori, T. Wandless, G. D. Shelton, and M. Montminy. 2007. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13597-603. - PubMed
- Bodine, S. C., E. Latres, S. Baumhueter, V. K. Lai, L. Nunez, B. A. Clarke, W. T. Poueymirou, F. J. Panaro, E. Na, K. Dharmarajan, Z. Q. Pan, D. M. Valenzuela, T. M. DeChiara, T. N. Stitt, G. D. Yancopoulos, and D. J. Glass. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2941704-1708. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous