Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival - PubMed (original) (raw)
Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival
Atsushi Sawada et al. Mol Cell Biol. 2008 May.
Abstract
Four members of the TEAD/TEF family of transcription factors are expressed widely in mouse embryos and adult tissues. Although in vitro studies have suggested various roles for TEAD proteins, their in vivo functions remain poorly understood. Here we examined the role of Tead genes by generating mouse mutants for Tead1 and Tead2. Tead2(-/-) mice appeared normal, but Tead1(-/-); Tead2(-/-) embryos died at embryonic day 9.5 (E9.5) with severe growth defects and morphological abnormalities. At E8.5, Tead1(-/-); Tead2(-/-) embryos were already small and lacked characteristic structures such as a closed neural tube, a notochord, and somites. Despite these overt abnormalities, differentiation and patterning of the neural plate and endoderm were relatively normal. In contrast, the paraxial mesoderm and lateral plate mesoderm were displaced laterally, and a differentiated notochord was not maintained. These abnormalities and defects in yolk sac vasculature organization resemble those of mutants for Yap, which encodes a coactivator of TEAD proteins. Moreover, we demonstrated genetic interactions between Tead1 and Tead2 and Yap. Finally, Tead1(-/-); Tead2(-/-) embryos showed reduced cell proliferation and increased apoptosis. These results suggest that Tead1 and Tead2 are functionally redundant, use YAP as a major coactivator, and support notochord maintenance as well as cell proliferation and survival in mouse development.
Figures
FIG. 1.
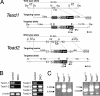
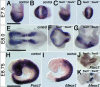
Targeted disruption of the Tead1 and Tead2 genes. (A) The Tead1 mutant allele was generated by replacing exon 2, containing the translation initiation site, with the _loxP_-_Pgk_-_neo_-poly(A)-loxP cassette (neo) and the SA-IRES-β-geo-pA signal cassette. The Tead2 mutant allele was generated by replacing exons 2 and 3, containing the translation initiation site and half of the TEA domain, with SA-IRES-β-geo-pA. See Materials and Methods for details. Positions of the 5′ and 3′ probes for Southern hybridization are indicated as 5′P and 3′P, respectively. Positions of the PCR primers used for the initial screening of homologous recombinants and genotype identifications are indicated by arrows and arrowheads, respectively. Abbreviations are as follows: Sw, SwaI; Ec, Eco47III; N, NaeI; Ag, AgeI; S, SalI; E, EagI; K, KpnI; A, AflIII; Nr, NruI; DT-A, MC1-DT-A-pA cassette. (B) Expression of Tead1 and Tead2 transcripts in the mutant embryos. RT-PCR was performed using RNAs isolated from E8.5 control or _Tead1_−/−; _Tead2_−/− (DKO) embryos and primer sets for various regions of Tead1 and Tead2 cDNAs. In the double mutant embryos, no Tead1 transcript was detected. Although some transcripts were detected for Tead2 (Tead2-2 and Tead2-3), these transcripts lacked coding regions for the TEA domain and coactivator-interacting domain (Tead2-1 and Tead2-4). These truncated Tead2 transcripts would not encode a functional protein. (C) Genotype determination by PCR. The genotypes of the embryos were clearly identified by PCR amplification of yolk sac genomic DNA fragments with a mixture of the three primers indicated by arrowheads in panel A (Tead1 locus: F1, F2, and R2; Tead2 locus: F2, S2, and R2).
FIG. 2.
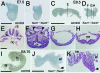
Gross phenotypes of the _Tead1_−/−; _Tead2_−/− mutant embryos. External views (A to D, I, and J) and histological sections (F to H and K) of control (A, C, E, and I) and _Tead1_−/−; _Tead2_−/− (B, D, F to H, J, and K) embryos. Anterior is to the left. Images shown in panels E and F to H are transverse sections of the images shown in panels C and D, respectively. The approximate positions of sections are indicated in panels C and D. Panel K shows a sagittal section of panel J. The _Tead1_−/−; _Tead2_−/− embryos were smaller than control embryos. Although the mutants were morphologically normal at E7.5 (B), their morphology was disorganized by E8.5 (D). Heart tubes failed to fuse (arrowheads in panel F). At E8.75, the neural plate undulated dorsoventrally, and an abnormal protrusion developed in the posterior-ventral region (arrowheads in panels J and K). Abbreviations: h, heart tubes; nc, notochord; ne, neuroectoderm; nt, neural tube; s, somite; al, allantois. Scale bars and use of the same scalfe in other panels are as follows: 400 μm (A; also panel B), 1 mm (C; also panel D), 100 μm (E; also panels F to H), 1 mm (I), and 500 μm (J; also panel K).
FIG. 3.
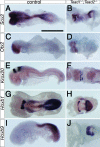
Specification and anteroposterior patterning of the neuroectoderm were relatively normal in _Tead1_−/−; _Tead2_−/− embryos. Control and _Tead1_−/−; _Tead2_−/− embryos were examined for expression of the pan-neural gene Sox2 (A and B) and regionally restricted genes Otx2 (C and D, fore- and midbrain), Krox20 (E and F, r3 and r5), Hoxb1 (G and H, r4 and posterior to the thoracic region) and Hoxb9 (I and J; posterior to the thoracic region). In spite of the gross morphological abnormalities, the mutant embryos retained an essentially proper spatial organization of gene expression patterns. Scale bar, 1 mm (same scale used for all panels).
FIG. 4.
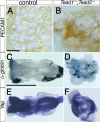
Endoderm development in the _Tead1_−/−; _Tead2_−/− embryo was relatively normal except for the posterior definitive endoderm. Control and _Tead1_−/−; _Tead2_−/− embryos were examined for the expression of a visceral endoderm-specific gene, Afp (A and B) and definitive endoderm genes, Foxa1 (C and D) and Sox17 (E and F). Specification of the visceral and definitive endoderm occurred relatively normally except for the posterior definitive endoderm region, in which the expression of Sox17 was reduced. Abbreviations: al, allantois; n, node; ps, primitive streak. Scale bar, 1 mm (same scale used for all panels).
FIG. 5.
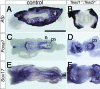
Paraxial mesoderm was physically displaced toward the lateral margins in _Tead1_−/−; _Tead2_−/− embryos. Control and _Tead1_−/−; _Tead2_−/− embryos were examined for expression of a paraxial mesoderm gene, Foxc1 (A to H and J) and a somite-specific gene, Meox1 (I and K). The Foxc1 expression domain was normal at E7.5, but it was displaced toward the lateral edges at E8.0 and E8.75. Foxc1 was also expressed in the anterior region of the mutants, but that expression domain is obscured by other tissue in panel J. Similar lateral expression was also observed for Meox1 at E8.75. Panels A, C, G, H, and I show lateral views; panels B, D, F, J, and K show ventral views; panel E shows a dorsal view. Scale bars and use of the same scale in other panels are as follows: 400 μm (A; also panels B to D), 750 μm (E; also panels F and G), and in 800 μm (H; also panels I to K).
FIG. 6.
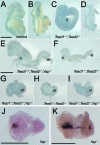
Axial mesoderm formed but was not maintained in _Tead1_−/−; _Tead2_−/− embryos. Expression of Foxa2 and Brachyury was examined in control and mutant embryos. Anterior is to the left. Foxa2 expression was detected in the midline tissue, including the prechordal plate, the floor plate of the neural tube/plate, the notochord, and the node (A). In the mutant embryos, Foxa2 expression was initially normal at E8.25 (B) but became discontinuous by E8.5 (C) and was lost except for the anterior tip at E8.75 (F). Similar changes in Brachyury expression in the node and notochord were observed (F to H). Scale bar, 1 mm (same scale used for all panels).
FIG. 7.
Vascular organization in the yolk sac was disorganized in _Tead1_−/−; _Tead2_−/− embryos. Expression of PECAM-1, α-globin, and Yap was examined in control (A, C, and E) and mutant (B, D, and F) embryos. PECAM-1-positive endothelial cells were differentiated in the mutant yolk sac, but they failed to form organized vasculature (B). Erythroblasts expressing α-globin also formed in the mutant yolk sac (D). Although the _Tead1_−/−; _Tead2_−/− embryos resembled Yap tm1/tm1 mutants, Yap expression in the _Tead1_−/−; _Tead2_−/− embryos was not significantly affected (E and F). Scale bars, 100 μm (A; same scale used in panel B) and 1 mm (C; same scale used in panels D to F).
FIG. 8.
Tead1 and Tead2 genetically interacted with Yap. (A to I) External morphologies of wild-type and Tead1; Tead2; Yap compound mutant embryos at E9.5. Half of the Tead1+/−; _Tead2_−/− embryos showed an open-brain phenotype (C and D), and the Tead1+/−; _Tead2_−/−; Yap+/tm1 embryos showed additional defects in posterior development and embryonic turning (E). The _Tead1_−/−; Tead2+/− embryos showed severe growth retardation (F), and _Tead1_−/−; Tead2+/−; Yap+/tm1 embryos showed severe morphological abnormalities (G), which resembled those of _Tead1_−/−; _Tead2_−/− embryos (H). The _Tead1_−/−; _Tead2_−/−; Yap+/tm1 embryos (I) were essentially the same as _Tead1_−/−; _Tead2_−/− embryos. The _Tead1_−/−; Tead2+/−; Yap+/tm1, _Tead1_−/−; _Tead2_−/− and _Tead1_−/−; _Tead2_−/−; Yap+/tm1 embryos all developed a characteristic protrusion at the posterior-ventral region (arrowheads in panels G, H, and I) and had a bulbous allantois. (J and K) Foxa2 expression in Yap tm1/tm1 mutant embryos. Expression of the TEAD1/TEAD2 target gene, Foxa2, was either strongly down-regulated except for the anterior tip (J) or discontinuous and gradually down-regulated at the posterior region (K) in the Yap tm1/tm1 embryos. Embryos shown in J and K are at E8.75 and E9.5, respectively. Abbreviations: h, heart; al, allantois. Scale bars, 1 mm (A and E; same scale used for panels A to I) and 600 nm (J and K).
FIG. 9.
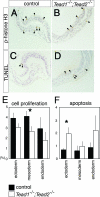
Cell proliferation and apoptosis were altered in _Tead1_−/−; _Tead2_−/− embryos. (A to D) Sections showing the distribution of proliferating (A and B) and apoptotic (C and D) cells (arrowheads) in the control and _Tead1_−/−; _Tead2_−/− embryos at E7.75. (E and F) Quantification of proliferating and apoptotic cells of control and Tead1_−/−; Tead2_−/− embryos detected by anti-phospho-histone H3 and TUNEL labeling, respectively. Values shown represent the means and standard errors (n = 5) of the percentages of total cells that were positive. Black and white bars represent control and _Tead1_−/−; _Tead2_−/− embryos, respectively. Asterisks above the bars indicate that the differences were statistically significant (P < 0.05).
Similar articles
- Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling.
Ota M, Sasaki H. Ota M, et al. Development. 2008 Dec;135(24):4059-69. doi: 10.1242/dev.027151. Epub 2008 Nov 12. Development. 2008. PMID: 19004856 - YAP/TAZ enhance mammalian embryonic neural stem cell characteristics in a Tead-dependent manner.
Han D, Byun SH, Park S, Kim J, Kim I, Ha S, Kwon M, Yoon K. Han D, et al. Biochem Biophys Res Commun. 2015 Feb 27;458(1):110-6. doi: 10.1016/j.bbrc.2015.01.077. Epub 2015 Jan 26. Biochem Biophys Res Commun. 2015. PMID: 25634692 - The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo.
Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE Jr. Weinstein DC, et al. Cell. 1994 Aug 26;78(4):575-88. doi: 10.1016/0092-8674(94)90523-1. Cell. 1994. PMID: 8069910 - An evolutionary, structural and functional overview of the mammalian TEAD1 and TEAD2 transcription factors.
Landin-Malt A, Benhaddou A, Zider A, Flagiello D. Landin-Malt A, et al. Gene. 2016 Oct 10;591(1):292-303. doi: 10.1016/j.gene.2016.07.028. Epub 2016 Jul 14. Gene. 2016. PMID: 27421669 Free PMC article. Review. - [T-Box genes and developmental decisions that cells make].
Korzh VP. Korzh VP. Ontogenez. 2001 May-Jun;32(3):196-203. Ontogenez. 2001. PMID: 11548409 Review. Russian.
Cited by
- TEAD transcription factor family emerges as a promising therapeutic target for oral squamous cell carcinoma.
Wang S, Shao D, Gao X, Zhao P, Kong F, Deng J, Yang L, Shang W, Sun Y, Fu Z. Wang S, et al. Front Immunol. 2024 Oct 4;15:1480701. doi: 10.3389/fimmu.2024.1480701. eCollection 2024. Front Immunol. 2024. PMID: 39430767 Free PMC article. Review. - Quit your YAPing: a new target for cancer therapy.
Stanger BZ. Stanger BZ. Genes Dev. 2012 Jun 15;26(12):1263-7. doi: 10.1101/gad.196501.112. Genes Dev. 2012. PMID: 22713867 Free PMC article. - How are pluripotent cells captured in culture?
Kinoshita M. Kinoshita M. Reprod Med Biol. 2015;14(3):85-98. doi: 10.1007/s12522-014-0199-8. Epub 2014 Dec 3. Reprod Med Biol. 2015. PMID: 26161037 Free PMC article. - The hippo signaling pathway in development and cancer.
Pan D. Pan D. Dev Cell. 2010 Oct 19;19(4):491-505. doi: 10.1016/j.devcel.2010.09.011. Dev Cell. 2010. PMID: 20951342 Free PMC article. Review. - Transcriptional Regulation of the Hippo Pathway: Current Understanding and Insights from Single-Cell Technologies.
Paul S, Xie S, Yao X, Dey A. Paul S, et al. Cells. 2022 Jul 17;11(14):2225. doi: 10.3390/cells11142225. Cells. 2022. PMID: 35883668 Free PMC article. Review.
References
- Abdelkhalek, H. B., A. Beckers, K. Schuster-Gossler, M. N. Pavlova, H. Burkhardt, H. Lickert, J. Rossant, R. Reinhardt, L. C. Schalkwyk, I. Muller, B. G. Herrmann, M. Ceolin, R. Rivera-Pomar, and A. Gossler. 2004. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 181725-1736. - PMC - PubMed
- Ang, S. L., and J. Rossant. 1994. HNF-3 β is essential for node and notochord formation in mouse development. Cell 78561-574. - PubMed
- Burke, A. C., C. E. Nelson, B. A. Morgan, and C. Tabin. 1995. Hox genes and the evolution of vertebrate axial morphology. Development 121333-346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases