Mammalian DNA methyltransferases: a structural perspective - PubMed (original) (raw)
Review
Mammalian DNA methyltransferases: a structural perspective
Xiaodong Cheng et al. Structure. 2008 Mar.
Abstract
The methylation of mammalian DNA, primarily at CpG dinucleotides, has long been recognized to play a major role in controlling gene expression, among other functions. Given their importance, it is surprising how many basic questions remain to be answered about the proteins responsible for this methylation and for coordination with the parallel chromatin-marking system that operates at the level of histone modification. This article reviews recent studies on, and discusses the resulting biochemical and structural insights into, the DNA nucleotide methyltransferase (Dnmt) proteins 1, 3a, 3a2, 3b, and 3L.
Figures
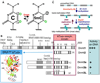
Figure 1. DNA Methylation
(A) DNA cytosine methylation at ring carbon C5. See the text for a summary of the mechanism. The question mark indicates possible activity of DNA demethylases (Kress et al., 2006; Vairapandi, 2004). (B) Members of the DNMT family. Schematic representation of Dnmt1 and Dnmt3. Dnmt2 is a tRNAAsp MTase (Goll et al., 2006) (insert). Roman numerals refer to conserved motifs of DNA MTases (Kumar et al., 1994); motif IV includes the Cys nucleophile that forms a transient covalent bond to C6 of the target cytosine. Other details are explained in the text or in work by Goll and Bestor (2005). (C) Maintenance versus de novo methylation. As described in the text, the roles of the Dnmts are not completely distinct in this respect. The pale-blue segments are substrate sequences (usually CpG), and the turquoise shapes represent methyl groups on the cytosines. After replication or repair, the duplex is methylated on one strand only.
Figure 2. Domain Structures of the Dnmt3 Family
(A) The PWWP domain structure of Dnmt3b, shown as a GRASP output (Nicholls et al., 1991), is rich in basic residues (Qiu et al., 2002). Selected charged, surface-exposed residues are indicated. (B) Structure of Dnmt3L with a bound histone H3 N-terminal tail (orange) (Ooi et al., 2007). (C) A surface representation of the Dnmt3a-C/3L-C tetramer, with two short DNA molecules adopted by superimposition of the HhaI-DNA complex structure (Klimasauskas et al., 1994) onto individual Dnmt3a-C molecules. The figure was adapted from work by Jia et al. (2007). (D) The Dnmt3a-C/3L-C tetramer with one contiguous curved DNA molecule covering two active sites. The figure was adapted from work by Jia et al. (2007). (E) The Dnmt3a dimer could, in theory, methylate two CpGs separated by one helical turn in one binding event.
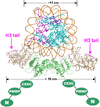
Figure 3. A Model for Interaction between a Dnmt3a-3L Tetramer and a Nucleosome
A nucleosome is shown, docked to a Dnmt3L-3a-3a-3L tetramer (3a-C in green; 3L full length in gray). The position of a peptide derived from the sequence of the histone H3 amino terminus (purple) is shown and is taken from a cocrystal structure with this peptide bound to Dnmt3L (Ooi et al., 2007). When the tetramer is wrapped around the nucleosome, the two Dnmt3L molecules could bind both histone tails from one nucleosome. The amino-proximal portion of Dnmt3a is shown in cartoon form, with domains labeled as N (for the variable region at the amino terminus), PWWP (which may be involved in nonspecific DNA binding), and CXXC (a Cys-rich 3-Zn-binding domain). By analogy to Dnmt3L, the CXXC domain of Dnmt3a might interact with histone tails from neighboring nucleosomes.
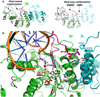
Figure 4. The 3a-3L Interface May Contribute to the Stability of Both AdoMet Binding and the Active-Site Loop Conformation
(A) Effects on the active-site loop. Left panel: superimposition of Dnmt3a and the protein component of the M.HhaI-DNA complex (PDB code: 1mht; gray). This superimposition has a root-mean-square deviation of 2.3 Å (comparing 220 pairs of Cα atoms by DALI) or 0.8 Å (manually aligning 53 pairs of Cα atoms forming the central 7 β strands). Right panel: superimposition of Dnmt3a and M.HhaI in the absence of DNA (PDB code: 1hmy). The M.HhaI active-site loop (gray) is shown from both structures: the closed, catalytically competent form adopted in the presence of DNA, and the open form adopted in its absence. (B) Effects on AdoMet binding. Dnmt3a-C is shown in green, Dnmt3L-C is shown in cyan, and the active-site loop of Dnmt3a is shown in magenta. AdoHcy (yellow) and residues involved in interactions are shown as stick models, and the corresponding ICF mutations in human Dnmt3b are shown in red (see Figure 6C). The panel was adopted from Supplemental Data in Jia et al. (2007).
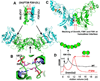
Figure 5. Structures of the 3a-3L and 3a-3a Interfaces
(A) Structure of the Dnmt3a-3L interface (PDB code: 2QRV). (B) Structure of the Dnmt3a-3a interface (PDB code: 2QRV). (C) Structure of the Dnmt3L-3L interface (PDB code: 2PV0). (D) Size-exclusion chromatography of wild-type Dnmt3a2 (black trace) or of an F728A mutant (red trace) (adopted from Supplemental Data in Jia et al. [2007]). The cartoons indicate the interpretation of oligomerization; the small yellow ovals indicate the Dnmt3a DNA-binding region (though it is absent altogether in Dnmt3L), and the other oval indicates Phe (dark green, WT) or Ala (red, mutant).
Figure 6. Sequences of the 3a-3L and 3a-3a Interfaces
(A) The Dnmt3a-3L interface is shown; secondary structural elements are indicated in light blue. Numbering above the sequences corresponds to the mouse ortholog. White-on-black residues are conserved in Dnmt3a, Dnmt3b, and Dnmt3L, whereas gray-highlighted positions are conserved (with ≤ 1 substitution) between 3a and 3b. The red highlighted C is the cysteine nucleophile of the MTase active site, whereas human Dnmt3b amino acids shown in red are associated with the disease ICF when altered in specific ways (see text). Positions highlighted in yellow are responsible for interactions between Dnmt3a and Dnmt3L (indicated by lines). The range of vertebrates with obvious Dnmt3L orthologs is much more restricted than that for Dnmt3a or Dnmt3b. (B) The Dnmt3a-3a interface is shown; the sequence is highlighted as in (A). The block arrows underneath indicate reciprocal salt bridges between the conserved Arg and Asp, as shown at Figure 5B. (C) Distribution of human ICF mutations on Dnmt3b (in parentheses). The panel was adopted from the Supplemental Data in Jia et al. (2007).
Similar articles
- Mammalian DNA methyltransferases.
Siedlecki P, Zielenkiewicz P. Siedlecki P, et al. Acta Biochim Pol. 2006;53(2):245-56. Epub 2006 Apr 3. Acta Biochim Pol. 2006. PMID: 16582985 Review. - Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation.
Papa CM, Springer NM, Muszynski MG, Meeley R, Kaeppler SM. Papa CM, et al. Plant Cell. 2001 Aug;13(8):1919-28. doi: 10.1105/tpc.010064. Plant Cell. 2001. PMID: 11487702 Free PMC article. - DNA methylation-mediated epigenetic control.
Rottach A, Leonhardt H, Spada F. Rottach A, et al. J Cell Biochem. 2009 Sep 1;108(1):43-51. doi: 10.1002/jcb.22253. J Cell Biochem. 2009. PMID: 19565567 - Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase.
Vilkaitis G, Suetake I, Klimasauskas S, Tajima S. Vilkaitis G, et al. J Biol Chem. 2005 Jan 7;280(1):64-72. doi: 10.1074/jbc.M411126200. Epub 2004 Oct 27. J Biol Chem. 2005. PMID: 15509558 - The DNA methyltransferases of mammals.
Bestor TH. Bestor TH. Hum Mol Genet. 2000 Oct;9(16):2395-402. doi: 10.1093/hmg/9.16.2395. Hum Mol Genet. 2000. PMID: 11005794 Review.
Cited by
- Structural insight into the DNMT1 reaction cycle by cryo-electron microscopy.
De I, Weidenhausen J, Concha N, Müller CW. De I, et al. PLoS One. 2024 Sep 3;19(9):e0307850. doi: 10.1371/journal.pone.0307850. eCollection 2024. PLoS One. 2024. PMID: 39226277 Free PMC article. - Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Shao Z, Lu J, Khudaverdyan N, Song J. Shao Z, et al. Nat Commun. 2024 Aug 9;15(1):6815. doi: 10.1038/s41467-024-51246-4. Nat Commun. 2024. PMID: 39122718 Free PMC article. - Evaluating the link between DIO3-FA27 promoter methylation, biochemical indices, and heart failure progression.
Qi Y, Meng X, Li J, He A, Hao J, Zhao X, Zhao R, Chen R, Zhang R. Qi Y, et al. Clin Epigenetics. 2024 Apr 24;16(1):57. doi: 10.1186/s13148-024-01668-0. Clin Epigenetics. 2024. PMID: 38659084 Free PMC article. - Consideration of SHP-1 as a Molecular Target for Tumor Therapy.
Lim S, Lee KW, Kim JY, Kim KD. Lim S, et al. Int J Mol Sci. 2023 Dec 26;25(1):331. doi: 10.3390/ijms25010331. Int J Mol Sci. 2023. PMID: 38203502 Free PMC article. Review. - Epigenetic regulation of craniofacial development and disease.
Shull LC, Artinger KB. Shull LC, et al. Birth Defects Res. 2024 Jan;116(1):e2271. doi: 10.1002/bdr2.2271. Epub 2023 Nov 14. Birth Defects Res. 2024. PMID: 37964651 Review.
References
- Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2007 in press. Published online October 15, 2007. - PubMed
- Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007.
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
- Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM068680/GM/NIGMS NIH HHS/United States
- R01 GM049245-15/GM/NIGMS NIH HHS/United States
- R01 GM068680/GM/NIGMS NIH HHS/United States
- R01 GM049245/GM/NIGMS NIH HHS/United States
- GM049245/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases