The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies - PubMed (original) (raw)
The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies
Alessandro Achilli et al. PLoS One. 2008.
Abstract
Only a limited number of complete mitochondrial genome sequences belonging to Native American haplogroups were available until recently, which left America as the continent with the least amount of information about sequence variation of entire mitochondrial DNAs. In this study, a comprehensive overview of all available complete mitochondrial DNA (mtDNA) genomes of the four pan-American haplogroups A2, B2, C1, and D1 is provided by revising the information scattered throughout GenBank and the literature, and adding 14 novel mtDNA sequences. The phylogenies of haplogroups A2, B2, C1, and D1 reveal a large number of sub-haplogroups but suggest that the ancestral Beringian population(s) contributed only six (successful) founder haplotypes to these haplogroups. The derived clades are overall starlike with coalescence times ranging from 18,000 to 21,000 years (with one exception) using the conventional calibration. The average of about 19,000 years somewhat contrasts with the corresponding lower age of about 13,500 years that was recently proposed by employing a different calibration and estimation approach. Our estimate indicates a human entry and spread of the pan-American haplogroups into the Americas right after the peak of the Last Glacial Maximum and comfortably agrees with the undisputed ages of the earliest Paleoindians in South America. In addition, the phylogenetic approach also indicates that the pathogenic status proposed for various mtDNA mutations, which actually define branches of Native American haplogroups, was based on insufficient grounds.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
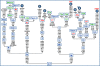
Figure 1. Basal tree encompassing the roots of Native American mtDNA haplogroups.
The tree is rooted on the haplogroup L3 founder and the position of the revised Cambridge reference sequence (rCRS) is indicated for reading off sequence motifs. Closely related Asian branches are indicated in green. Detailed phylogenies for the four pan-American haplogroups (A2, B2, C1, and D1, highlighted in red) are shown in the corresponding figures. The complete sequences that are currently available for the other four Native American haplogroups (X2a, C4c, D2a, and D4h3, highlighted in red) are also displayed. Haplogroup D3 is common among Inuit populations , but all complete sequences available are from Siberia , . As for A2a, the HVS-I motif (16111 16192 16223 16233 16290 16319 16331) of the reported sequence (no. 1) is common in Na-Dené groups . Sequence no. 2 has been revised taking into account that the originally reported transitions at 4732 and 5147 were artifacts due to a sample mix-up, while sequence no. 6 represents the shared motif of six Aleutian mitochondrial genomes . Mutations are transitions unless specified: suffixes indicate transversions (to A, G, C, or T) or indels (+, d). Mutations back to the rCRS nucleotide are prefixed with @. Recurrent mutational events are underlined. Mutations in italics are either disease-causing or heteroplasmic or likely erroneous (and do not enter age calculations). We have followed the recent guidelines for standardization of the alignment in long C stretches , but disregarded any length variation in the C stretches that would then be scored at 309 or 16193 (which is often subject to considerable heteroplasmy). A number flagging a circled haplotype indicates the number of individuals sharing the corresponding haplotype (if >1). Additional information is provided in Text S4, while Table S1 lists the source of the complete genomes.
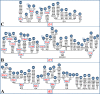
Figure 2. Phylogeny of complete mtDNA sequences belonging to haplogroup A2.
The sequencing procedure for the novel complete sequences and the phylogeny construction were performed as described elsewhere . Recurrent mutational events within the haplogroup are underlined, while mutations in italics are either disease-causing or heteroplasmic or likely erroneous, and were not used for age calculations. Table S1 lists the source of the complete genomes. For additional information, see the legend for Figure 1.
Figure 3. Phylogeny of complete mtDNA sequences belonging to haplogroups B2 (A), C1 (B) and D1 (C).
For additional information, see the legends for Figures 1 and 2.
Similar articles
- Large scale mitochondrial sequencing in Mexican Americans suggests a reappraisal of Native American origins.
Kumar S, Bellis C, Zlojutro M, Melton PE, Blangero J, Curran JE. Kumar S, et al. BMC Evol Biol. 2011 Oct 7;11:293. doi: 10.1186/1471-2148-11-293. BMC Evol Biol. 2011. PMID: 21978175 Free PMC article. - Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas.
Fagundes NJ, Kanitz R, Eckert R, Valls AC, Bogo MR, Salzano FM, Smith DG, Silva WA Jr, Zago MA, Ribeiro-dos-Santos AK, Santos SE, Petzl-Erler ML, Bonatto SL. Fagundes NJ, et al. Am J Hum Genet. 2008 Mar;82(3):583-92. doi: 10.1016/j.ajhg.2007.11.013. Epub 2008 Feb 28. Am J Hum Genet. 2008. PMID: 18313026 Free PMC article. - Identification of Whole Mitochondrial Genomes from Venezuela and Implications on Regional Phylogenies in South America.
Lee EJ, Merriwether DA. Lee EJ, et al. Hum Biol. 2015 Jan;87(1):29-38. doi: 10.13110/humanbiology.87.1.0029. Hum Biol. 2015. PMID: 26416320 - The initial peopling of the Americas: a growing number of founding mitochondrial genomes from Beringia.
Perego UA, Angerhofer N, Pala M, Olivieri A, Lancioni H, Hooshiar Kashani B, Carossa V, Ekins JE, Gómez-Carballa A, Huber G, Zimmermann B, Corach D, Babudri N, Panara F, Myres NM, Parson W, Semino O, Salas A, Woodward SR, Achilli A, Torroni A. Perego UA, et al. Genome Res. 2010 Sep;20(9):1174-9. doi: 10.1101/gr.109231.110. Epub 2010 Jun 29. Genome Res. 2010. PMID: 20587512 Free PMC article. - Ancient DNA perspectives on American colonization and population history.
Raff JA, Bolnick DA, Tackney J, O'Rourke DH. Raff JA, et al. Am J Phys Anthropol. 2011 Dec;146(4):503-14. doi: 10.1002/ajpa.21594. Epub 2011 Sep 13. Am J Phys Anthropol. 2011. PMID: 21913177 Review.
Cited by
- A reevaluation of the Native American mtDNA genome diversity and its bearing on the models of early colonization of Beringia.
Fagundes NJ, Kanitz R, Bonatto SL. Fagundes NJ, et al. PLoS One. 2008 Sep 17;3(9):e3157. doi: 10.1371/journal.pone.0003157. PLoS One. 2008. PMID: 18797501 Free PMC article. - Pan-American mDNA haplogroups in Chilean patients with Leber's hereditary optic neuropathy.
Romero P, Fernández V, Slabaugh M, Seleme N, Reyes N, Gallardo P, Herrera L, Peña L, Pezo P, Moraga M. Romero P, et al. Mol Vis. 2014 Mar 14;20:334-40. eCollection 2014. Mol Vis. 2014. PMID: 24672219 Free PMC article. - Biogeographical origin and timing of the founder ichthyosis TGM1 c.1187G > A mutation in an isolated Ecuadorian population.
Esperón-Moldes US, Pardo-Seco J, Montalván-Suárez M, Fachal L, Ginarte M, Rodríguez-Pazos L, Gómez-Carballa A, Moscoso F, Ugalde-Noritz N, Ordóñez-Ugalde A, Tettamanti-Miranda D, Ruiz JC, Salas A, Vega A. Esperón-Moldes US, et al. Sci Rep. 2019 May 9;9(1):7175. doi: 10.1038/s41598-019-43133-6. Sci Rep. 2019. PMID: 31073126 Free PMC article. - Mitochondrial DNA control region diversity in a population from Parana state-increasing the Brazilian forensic database.
Poletto MM, Malaghini M, Silva JS, Bicalho MG, Braun-Prado K. Poletto MM, et al. Int J Legal Med. 2019 Mar;133(2):347-351. doi: 10.1007/s00414-018-1886-5. Epub 2018 Jun 29. Int J Legal Med. 2019. PMID: 29959556 - MtDNA ancestry of Rio de Janeiro population, Brazil.
Bernardo S, Hermida R, Desidério M, Silva DA, de Carvalho EF. Bernardo S, et al. Mol Biol Rep. 2014;41(4):1945-50. doi: 10.1007/s11033-014-3041-9. Epub 2014 Jan 14. Mol Biol Rep. 2014. PMID: 24420852
References
- Schurr TG, Sherry ST. Mitochondrial DNA and Y chromosome diversity and the peopling of the Americas: evolutionary and demographic evidence. Am J Hum Biol. 2004;16:420–439. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources