Comparative lesion sequencing provides insights into tumor evolution - PubMed (original) (raw)
Comparative Study
. 2008 Mar 18;105(11):4283-8.
doi: 10.1073/pnas.0712345105. Epub 2008 Mar 12.
Wei-Dong Chen, Giovanni Parmigiani, Frank Diehl, Niko Beerenwinkel, Tibor Antal, Arne Traulsen, Martin A Nowak, Christopher Siegel, Victor E Velculescu, Kenneth W Kinzler, Bert Vogelstein, Joseph Willis, Sanford D Markowitz
Affiliations
- PMID: 18337506
- PMCID: PMC2393770
- DOI: 10.1073/pnas.0712345105
Comparative Study
Comparative lesion sequencing provides insights into tumor evolution
Siân Jones et al. Proc Natl Acad Sci U S A. 2008.
Abstract
We show that the times separating the birth of benign, invasive, and metastatic tumor cells can be determined by analysis of the mutations they have in common. When combined with prior clinical observations, these analyses suggest the following general conclusions about colorectal tumorigenesis: (i) It takes approximately 17 years for a large benign tumor to evolve into an advanced cancer but <2 years for cells within that cancer to acquire the ability to metastasize; (ii) it requires few, if any, selective events to transform a highly invasive cancer cell into one with the capacity to metastasize; (iii) the process of cell culture ex vivo does not introduce new clonal mutations into colorectal tumor cell populations; and (iv) the rates at which point mutations develop in advanced cancers are similar to those of normal cells. These results have important implications for understanding human tumor pathogenesis, particularly those associated with metastasis.
Conflict of interest statement
Conflict of interest statement: Under separate licensing agreements between The Johns Hopkins University and Exact Sciences Corporation and Genzyme Corporation Oncology, V.E.V., K.W.K., and B.V are entitled to a share of royalty received by the University on sales of products described in this article/presentation. V.E.V., K.W.K., and B.V and the University own Genzyme Molecular Oncology stock, which is subject to certain restrictions under University policy. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
Figures
Fig. 1.
Major genetic alterations associated with colorectal tumorigenesis. See
SI Methods
for further explanation.
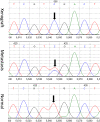
Fig. 2.
Representative examples of sequencing chromatograms of DNA from a xenograft, from the metastatic lesion from which the xenograft was derived, and from the patient's normal cells. Note that the ratio of the mutant to wild-type allele in the xenograft is higher than that in the metastatic lesion because the latter represented a mixture of neoplastic and nonneoplastic cells (stroma, white blood cells, etc.). The arrow points to the mutated base.
Fig. 3.
Histopathology of representative lesions. (A) Primary invasive moderately differentiated adenocarcinoma (enclosed by black boundary) arising in a tubular adenoma (enclosed by red boundary) from patient 10. (B) Primary invasive moderately differentiated adenocarcinoma (enclosed by black boundary) with adjacent nonneoplastic colonic mucosa (enclosed by red boundary) from patient 2. (C) Metastatic adenocarcinoma (enclosed by black boundary) to liver (enclosed by red boundary) derived from primary colon adenocarcinoma of patient 2. All sections were stained with H&E, and the tissues within each boundary were separately microdissected.
Fig. 4.
Representative examples of BEAMing assays from the indicated patients and lesions. In patient 13, the mutation shown represents one that was present in a new metastasis that occurred 29 months after chemotherapy (see Application to Individual Patients). The red dots correspond to beads attached to mutant DNA fragments [labeled with phycoerythrin (PE)], the blue dots correspond to beads attached to WT DNA fragments [labeled with fluorescein (FITC)], and the black dots correspond to beads attached to both WT and mutant DNA fragments.
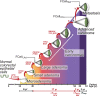
Fig. 5.
Evolution of a lethal cancer. Each cell-filled cone represents one or more clonal expansions (see
SI Methods
for details). The times required for the evolution of the large adenoma founder cell to an advanced carcinoma founder cell (ΔLAd,ACa) and evolution of the advanced carcinoma founder cell to metastatic founder cell (ΔACa,Met) were determined by comparative lesion sequencing. Other intervals, such as the time (_T_exp) required for the expansion of the metastasis founder cell FCellMet to the size detected in our patients, were estimated as described in
SI Methods
. The model posits that there are at least two clonal expansions, denoted by question marks, that are not associated with any known genetic alterations.
Similar articles
- Role of anti-oncomirs miR-143 and -145 in human colorectal tumors.
Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, Nakashima R, Kitade Y, Naoe T. Akao Y, et al. Cancer Gene Ther. 2010 Jun;17(6):398-408. doi: 10.1038/cgt.2009.88. Epub 2010 Jan 22. Cancer Gene Ther. 2010. PMID: 20094072 - Computational Identification of Novel Stage-Specific Biomarkers in Colorectal Cancer Progression.
Palaniappan A, Ramar K, Ramalingam S. Palaniappan A, et al. PLoS One. 2016 May 31;11(5):e0156665. doi: 10.1371/journal.pone.0156665. eCollection 2016. PLoS One. 2016. PMID: 27243824 Free PMC article. - Tristetraprolin regulates expression of VEGF and tumorigenesis in human colon cancer.
Lee HH, Son YJ, Lee WH, Park YW, Chae SW, Cho WJ, Kim YM, Choi HJ, Choi DH, Jung SW, Min YJ, Park SE, Lee BJ, Cha HJ, Park JW. Lee HH, et al. Int J Cancer. 2010 Apr 15;126(8):1817-1827. doi: 10.1002/ijc.24847. Int J Cancer. 2010. PMID: 19697322 - Colon Tumors with the Simultaneous Induction of Driver Mutations in APC, KRAS, and PIK3CA Still Progress through the Adenoma-to-carcinoma Sequence.
Hadac JN, Leystra AA, Paul Olson TJ, Maher ME, Payne SN, Yueh AE, Schwartz AR, Albrecht DM, Clipson L, Pasch CA, Matkowskyj KA, Halberg RB, Deming DA. Hadac JN, et al. Cancer Prev Res (Phila). 2015 Oct;8(10):952-61. doi: 10.1158/1940-6207.CAPR-15-0003. Epub 2015 Aug 14. Cancer Prev Res (Phila). 2015. PMID: 26276752 Free PMC article. - Autocrine induction of invasion and metastasis by tumor-associated trypsin inhibitor in human colon cancer cells.
Gouyer V, Fontaine D, Dumont P, de Wever O, Fontayne-Devaud H, Leteurtre E, Truant S, Delacour D, Drobecq H, Kerckaert JP, de Launoit Y, Bracke M, Gespach C, Desseyn JL, Huet G. Gouyer V, et al. Oncogene. 2008 Jul 3;27(29):4024-33. doi: 10.1038/onc.2008.42. Epub 2008 Mar 3. Oncogene. 2008. PMID: 18317448 Review.
Cited by
- Selection of optimal extraction and RT-PCR protocols for stool RNA detection of colorectal cancer associated immune genes.
Omran TA, Madsø IL, Sæther PC, Bemanian V, Tunsjø HS. Omran TA, et al. Sci Rep. 2024 Nov 10;14(1):27468. doi: 10.1038/s41598-024-78680-0. Sci Rep. 2024. PMID: 39523395 Free PMC article. - Spatiotemporal lineage tracing reveals the dynamic spatial architecture of tumor growth and metastasis.
Jones MG, Sun D, Min KHJ, Colgan WN, Tian L, Weir JA, Chen VZ, Koblan LW, Yost KE, Mathey-Andrews N, Russell AJC, Stickels RR, Balderrama KS, Rideout WM 3rd, Chang HY, Jacks T, Chen F, Weissman JS, Yosef N, Yang D. Jones MG, et al. bioRxiv [Preprint]. 2024 Oct 24:2024.10.21.619529. doi: 10.1101/2024.10.21.619529. bioRxiv. 2024. PMID: 39484491 Free PMC article. Preprint. - Improved diagnostic efficiency of CRC subgroups revealed using machine learning based on intestinal microbes.
Liu G, Su L, Kong C, Huang L, Zhu X, Zhang X, Ma Y, Wang J. Liu G, et al. BMC Gastroenterol. 2024 Sep 17;24(1):315. doi: 10.1186/s12876-024-03408-3. BMC Gastroenterol. 2024. PMID: 39289618 Free PMC article. - Simultaneous Detection of Collagen I Alpha II and Cytokeratin 19 mRNA by Multiplex qPCR in Liquid Biopsy in Diagnosis of Patients with Resectable Solid Tumors.
Estévez Pérez LS, Alén BO, Otero Alén M, Hormaetxe SD, Simón L, Concha Á. Estévez Pérez LS, et al. Int J Mol Sci. 2024 Sep 3;25(17):9567. doi: 10.3390/ijms25179567. Int J Mol Sci. 2024. PMID: 39273514 Free PMC article. - Cancer metastases: Tailoring the targets.
Pote MS, Singh D, M A A, Suchita J, Gacche RN. Pote MS, et al. Heliyon. 2024 Aug 2;10(15):e35369. doi: 10.1016/j.heliyon.2024.e35369. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170575 Free PMC article. Review.
References
- Kinzler KW, Vogelstein B. Lessons from hereditary colon cancer. Cell. 1996;87:159–170. - PubMed
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. - PubMed
- Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. - PubMed
- Markowitz S, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. - PubMed
- Thiagalingam S. Evaluation of chromosome 18q in colorectal cancers. Nat Genet. 1996;13:343–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA127306/CA/NCI NIH HHS/United States
- CA62924/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM078986-02/GM/NIGMS NIH HHS/United States
- CA043703/CA/NCI NIH HHS/United States
- CA116867/CA/NCI NIH HHS/United States
- R37 CA057345/CA/NCI NIH HHS/United States
- R01 CA057345/CA/NCI NIH HHS/United States
- R01 CA105090/CA/NCI NIH HHS/United States
- CA57345/CA/NCI NIH HHS/United States
- R37 CA043460/CA/NCI NIH HHS/United States
- CA121113/CA/NCI NIH HHS/United States
- GM078986/GM/NIGMS NIH HHS/United States
- CA127306/CA/NCI NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- R01 GM078986/GM/NIGMS NIH HHS/United States
- R01 CA120237/CA/NCI NIH HHS/United States
- R01 CA121113/CA/NCI NIH HHS/United States
- P30 CA043703/CA/NCI NIH HHS/United States
- CA43460/CA/NCI NIH HHS/United States
- CA120237/CA/NCI NIH HHS/United States
- U54 CA116867/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources