X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations - PubMed (original) (raw)
X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations
Yin Shen et al. Proc Natl Acad Sci U S A. 2008.
Abstract
X chromosome inactivation (XCI) is an essential mechanism for dosage compensation of X-linked genes in female cells. We report that subcultures from lines of female human embryonic stem cells (hESCs) exhibit variation (0-100%) for XCI markers, including XIST RNA expression and enrichment of histone H3 lysine 27 trimethylation (H3K27me3) on the inactive X chromosome (Xi). Surprisingly, regardless of the presence or absence of XCI markers in different cultures, all female hESCs we examined (H7, H9, and HSF6 cells) exhibit a monoallelic expression pattern for a majority of X-linked genes. Our results suggest that these established female hESCs have already completed XCI during the process of derivation and/or propagation, and the XCI pattern of lines we investigated is already not random. Moreover, XIST gene expression in subsets of cultured female hESCs is unstable and subject to stable epigenetic silencing by DNA methylation. In the absence of XIST expression, approximately 12% of X-linked promoter CpG islands become hypomethylated and a portion of X-linked alleles on the Xi are reactivated. Because alterations in dosage compensation of X-linked genes could impair somatic cell function, we propose that XCI status should be routinely checked in subcultures of female hESCs, with cultures displaying XCI markers better suited for use in regenerative medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
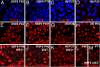
Fig. 1.
Different subcultures of hESCs (HSF6 and H9) exhibit varied XCI status. (A–D) XIST RNA FISH signal (red) shows XIST RNA coating on the Xi. Immunostaining of hESCs with antibodies against H3K27me3 (red) (E–H), H4K20me1 (red) (I and J), and macroH2A1 (red) (K–M). Punctate XIST FISH signals and foci of H3K27me3, H4K20me1, and macroH2A1 stainings indicate the presence of an Xi. Please note that, for _XIST_− hESCs, the punctate staining pattern of H4K20me1 in some hESCs cannot be seen because of overexposure of the image to compensate for the weakly stained cells.
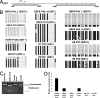
Fig. 2.
DNA methyation analysis of the XIST promoter and real-time RT-PCR analysis of XIST RNA levels. (A) Schematic diagram of XIST promoter/first exon and a further downstream region analyzed. CpG sites are presented in vertical bars, and the arrow indicates the transcription initiation site of XIST. CpG sites analyzed are underlined by black bars. (B) Bisulfite sequencing analysis reveals methylation patterns of promoter/first exon and a further downstream region of XIST gene in _XIST_+ and _XIST_− H9 and HSF6 hESCs and _XIST_− H7 hESCs in various passages. Each filled black dot represents one methylated CpG site, and an open dot represents an unmethylated CpG. (C) COBRA assay showing the XIST promoter for _XIST_− H9 and HSF6 are fully methylated, whereas the XIST promoter of _XIST_+ HSF6 is hypomethylated compared with normal female brain DNA. (D) Real-time quantitative PCR showing relative XIST expression levels in various samples of HSF6, H9, and H7 hESCs.
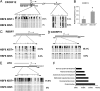
Fig. 3.
Analysis of methylation levels at promoter CpG islands in female hESCs in the presence or absence of XIST expression. (A) Bisulfite genomic sequencing analysis of the CpG island promoter and an exon region of CXORF12 gene in _XIST_+ and _XIST_− HSF6 cells. Note the promoter is 50% methylated in _XIST_+ HSF6 cells and becomes unmethylated in _XIST_− cells. In contrast, the exon region is 100% methylated in both _XIST_+ and _XIST_− cells. (B) Real-time quantitative PCR showing that the expression level of CXORF12 is significantly higher in _XIST_+ HSF6 cells compared with _XIST_− cells. *, P < 0.01. (C–E) Bisulfite methylation analysis in CpG islands of RBBP7, UTX, and CXORF15 genes. (F) Gene ontology analysis of 51 X-linked genes with decreased methylation levels in promoter CpG islands in _XIST_− hESCs (P < 0.05).
Fig. 4.
Relative gene expression levels of a subset of X-linked genes using pairs of HSF6 and H9 cells with or without XCI markers. (A) Real-time RT-PCR demonstrates increased mRNAs in HSF6 cells without XCI markers. Six X-linked genes including CXORF15, UTX, RBBP7, PLS3, SMARCA1, and PCTK1 were analyzed. (B) Real-time RT-PCR results of the same six genes for a pair of _XIST_+ and _XIST_− H9 cells. *, P < 0.01. (C) Real-time RT-PCR results of the RBBP7 and PLS6 for male hESCs (H1 and HSF1) and _XIST_+ and _XIST_− female hESCs (H9 and HSF6) cells. *, P < 0.05. (D) Models of dynamic regulation of XCI in female hESCs. Xa is shown in full-length X chromosome. Xi is depicted in an oval shape. Dotted X chromosome indicates partial reactivation. Paternal (p) and maternal (m) X are shown in blue and red.
References
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. - PubMed
- Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. - PubMed
- Maitra A, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. - PubMed
- Allegrucci C, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. - PubMed
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1F31MH070204/MH/NIMH NIH HHS/United States
- NS051411/NS/NINDS NIH HHS/United States
- F31 MH070204/MH/NIMH NIH HHS/United States
- R01 NS051411/NS/NINDS NIH HHS/United States
- R56 NS051411/NS/NINDS NIH HHS/United States
- R01 NS044405/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases