Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome - PubMed (original) (raw)
. 2008 Mar 15;68(6):1786-96.
doi: 10.1158/0008-5472.CAN-07-5547.
Aedín C Culhane, Michael W Y Chan, Chinnambally R Venkataramu, Mathias Ehrich, Aejaz Nasir, Benjamin A T Rodriguez, Joseph Liu, Pearlly S Yan, John Quackenbush, Kenneth P Nephew, Timothy J Yeatman, Tim H-M Huang
Affiliations
- PMID: 18339859
- PMCID: PMC4172329
- DOI: 10.1158/0008-5472.CAN-07-5547
Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome
Alfred S L Cheng et al. Cancer Res. 2008.
Abstract
Estrogen imprinting is used to describe a phenomenon in which early developmental exposure to endocrine disruptors increases breast cancer risk later in adult life. We propose that long-lived, self-regenerating stem and progenitor cells are more susceptible to the exposure injury than terminally differentiated epithelial cells in the breast duct. Mammospheres, containing enriched breast progenitors, were used as an exposure system to simulate this imprinting phenomenon in vitro. Using MeDIP-chip, a methylation microarray screening method, we found that 0.5% (120 loci) of human CpG islands were hypermethylated in epithelial cells derived from estrogen-exposed progenitors compared with the non-estrogen-exposed control cells. This epigenetic event may lead to progressive silencing of tumor suppressor genes, including RUNX3, in these epithelial cells, which also occurred in primary breast tumors. Furthermore, normal tissue in close proximity to the tumor site also displayed RUNX3 hypermethylation, suggesting that this aberrant event occurs in early breast carcinogenesis. The high prevalence of estrogen-induced epigenetic changes in primary tumors and the surrounding histologically normal tissues provides the first empirical link between estrogen injury of breast stem/progenitor cells and carcinogenesis. This finding also offers a mechanistic explanation as to why a tumor suppressor gene, such as RUNX3, can be heritably silenced by epigenetic mechanisms in breast cancer.
Figures
Figure 1
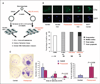
Preexposure of breast progenitor cells to estrogen increases proliferation of progeny epithelial cells. A, the breast progenitor cells were propagated as floating spherical colonies, called mammospheres, and treated with 70 nmol/L E2 or DMSO solvent control for 2 wk. To induce differentiation, cells were seeded on a collagen substratum in the absence of E2 for 2 to 3 wk. Phenotypic and epigenetic analyses were then performed on the progeny epithelial cells. B, immunofluorescence shows that some progenitor-derived epithelial cells express ERα. After E2 stimulation (10 nmol/L, 3 h), translocation of ERα protein from the cytoplasm and into the nucleus was seen, suggesting functional estrogen signaling. In contrast, epithelial cells preexposed to high-dose estrogen exhibited nuclear ERα localization before E2 stimulation, reminiscent of MCF-7 cells. The percentage, subcellular localization, and staining intensity of ERα-positive cells are shown in the bar chart. The numbers of cells listed in each category were independently scored by two researchers. C, left, cell proliferation was measured by colony formation assay. Representative results of two independent experiments are shown. Exposure of progenitor cells to estrogen significantly increased the number of large-sized colonies (P = 0.02; middle) and the overall size of colonies (P = 0.038; right) formed by epithelial progeny cells. Columns, mean of two independent experiments; bars, SD. Asterisks, statistical significance.
Figure 2
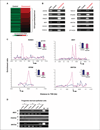
Preexposure of progenitor-derived epithelial cells to estrogen causes DNA hypermethylation and transcriptional silencing of tumor suppressor genes. A, differentially methylated genes in estrogen-preexposed progenitor-derived epithelial cells were identified by MeDIP-chip using a microarray panel containing 27,800 CpG islands. The maximum MeDIP enrichment along 120 hypermethylated loci in control and preexposed cells are shown in the heat map (see Supplementary Table S1 for a complete list of the hypermethylated loci). The heat map represents the averaged values of two independent experiments. B, to confirm candidate methylated genes determined by microarray, PCR primers targeting the MeDIP-enriched region were designed and the immunoprecipitated control/preexposed DNA samples were amplified along with input DNA. PCRs of no template (H2O) control are also shown. Twelve of 13 tested genes were validated by PCR analyses. C, DNA methylation landscaping maps by plotting the enrichment ratio of each probe of the hypermethylated genes. The preexposed cells (red line) displayed an enriched methylated region of multiple probes, within the CpG island, located in proximity to the TSS (arrow); no significant enrichment was observed in control cells (blue line). Genes showing DNA hypermethylation in preexposed cells were associated with transcriptional suppression [shown by real-time quantitative RT-PCR (qRT-PCR); _upper right, inset_]. Columns, mean of three independent experiments; bars, SD. D, methylation status of tumor suppressor gene promoter regions was confirmed by MSP in control/preexposed DNA samples derived from three individuals. MSP PCR primers targeting MeDIP-enriched regions were designed. Preexposed cells of at least one set of samples showed DNA methylation in these tumor suppressor genes. M, PCR product with primers specific for methylated genes; U, unmethylated genes.
Figure 3
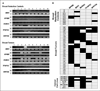
Estrogen-induced hypermethylation of tumor suppressor loci is also present in breast tumors. A, MSP analyses for tumor suppressor gene promoter methylation in breast reduction normal tissues (top) and primary tumors (bottom). Representative gel pictures show data for 10 of a total of 31 tumor tissues examined and 10 breast reduction normal tissues. B, summary of methylation data for all tissue samples and cancer cell lines. Aberrant methylation of these tumor suppressor genes occurred in a high percentage of primary tumors (23–74%) and cancer cell lines (25–100%) compared with reduction normal controls (0–20%).
Figure 4
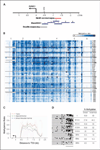
DNA hypermethylation of RUNX3 in breast tumors and adjacent histologically normal tissues. A, genomic map of the RUNX3 promoter CpG island. Arrow, RUNX3 TSS in the genome sequence; inverted arrow, potential estrogen-responsive element (ERE; predicted by MATCH using the TRANSFAC database). Red bar, MeDIP-enriched region. Locations of the RUNX3 promoter fragments interrogated by MassARRAY (blue bars) and bisulfite sequencing (gray bar). B, quantitative methylation results from MassARRAY analysis of RUNX3 promoter region in primary breast tissues. Six amplicons covering a total of 101 CpG sites, located approximately 0.3 to 2.3 kb upstream of the RUNX3 TSS, were designed to analyze 9 sets of breast tumor (T)/adjacent normal (N) tissues and 8 breast reduction normal tissues (BRN; 48 samples total). CpG site measurements in the analyzed region of the RUNX3 promoter for all samples are depicted in the heat map. Blue color intensity, methylation ratios (see scale bar); gray, missing data values. Each row represents methylation data from one tissue sample. Tissues from each cancer patient are separated by black lines. Each column (spaced by white lines) represents methylation data from each amplicon. C, summation of the overall methylation levels of each tissue type in a genomic context. Red dotted lines, methylation ratios of tumor tissues; green dotted lines, methylation ratios of adjacent normal tissues; blue dotted lines, methylation ratios of breast reduction normal tissues; gray lines, CpG content in the amplification region calculated for a moving 100-bp window. D, bisulfite sequencing analysis of RUNX3 promoter region in one set of breast tumor/adjacent normal tissues and three breast reduction normal tissues. Black dots, methylated CpG sites; white dots, unmethylated sites. The underlined region is near the beginning of the methylation plateau observed by MassARRAY. The percentages of CpG methylation of the overall and underlined regions are indicated for each tissue. Breast tumor and adjacent histologically normal tissues showed significantly higher methylation levels compared with breast reduction normal tissues, in agreement with the MassARRAY analysis. Additional bisulfite sequencing results are shown in Supplementary Fig. S3.
Figure 5
Mapping geographic zones of RUNX3 DNA methylation in histologically normal tissues adjacent to breast tumor. A, microdissected breast tissues from patients undergoing mastectomy were collected. Mastectomy specimens were marked for tumor and its surrounding zones N1 (1 cm), N2 (2 cm), N3 (3 cm), and N4 (4 cm) from the grossly visible tumor boundary. In bilateral and prophylactic mastectomy cases, normal breast tissues were taken from the four quadrants of the breast. UO, upper outer; Ul, upper inner; LO, lower outer; LI, lower inner. B, the graphs show methylation ratios of the primary breast tumor and adjacent normal tissues of indicated distance (cm) from tumor boundary of each patient. Data of contralateral (CL) prophylactic mastectomy normal epithelia of patient 10486 are also shown. Dotted lines, mean methylation ratio for breast reduction normal tissues as reference.
Figure 6
Inverse correlation between RUNX3 expression and ERα status. A, RUNX3 mRNA expression was significantly lower in IDC tissues compared with breast reduction normal tissues. No difference in RUNX3 expression was observed in adjacent normal tissues with unremarkable breast ducts and fibrocystic/nonproliferative changes versus reduction normal tissues. B, ERα-positive IDC had significantly reduced RUNX3 mRNA expression compared with ERα-negative tumors. Notably, normal breast tissues adjacent to ERα-positive breast tumors also had significantly lower expression than ERα-negative counterparts. C, evaluation of RUNX3 mRNA expression in a panel of breast cancer cell lines. ERα-positive cell lines had significantly lower RUNX3 expression than ERα-negative cell lines, in agreement with the data from primary breast tissues. In A to C, expression levels are represented in log2 values. In the box plots, the upper and lower boundaries of the box indicate 75th and 25th percentiles, respectively. Line within the box, median; bars above and below the box, 90th and 10th percentiles, respectively. The numbers of tissues/cell lines studied are also indicated. Asterisks, statistical significance. D, real-time quantitative RT-PCR analysis of RUNX3 in MCF-7 cells after a 5-d treatment with 5 µmol/L 5-aza-dC. Black line, 5-aza-dC-treated MCF-7 cells were hormone deprived for 72 h and treated with vehicle or 10 nmol/L E2 for the indicated times (h); gray line, 5-aza-dC-treated and hormone-deprived cells were treated with 4-OHT (1 µmol/L for 48 h) and then treated with vehicle or 10 nmol/L E2 for the indicated times (h). Results are presented relative to no 4-OHT treatment control (black line, 0 time point). Points, mean of triplicates; bars, SD.
Similar articles
- Tumor suppressor function of RUNX3 in breast cancer.
Chen LF. Chen LF. J Cell Biochem. 2012 May;113(5):1470-7. doi: 10.1002/jcb.24074. J Cell Biochem. 2012. PMID: 22275124 Free PMC article. - Enrichment of CpG island shore region hypermethylation in epigenetic breast field cancerization.
Muse ME, Titus AJ, Salas LA, Wilkins OM, Mullen C, Gregory KJ, Schneider SS, Crisi GM, Jawale RM, Otis CN, Christensen BC, Arcaro KF. Muse ME, et al. Epigenetics. 2020 Oct;15(10):1093-1106. doi: 10.1080/15592294.2020.1747748. Epub 2020 Apr 7. Epigenetics. 2020. PMID: 32255732 Free PMC article. - Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells.
Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng AS, Huang TH. Hsu PY, et al. Cancer Res. 2009 Jul 15;69(14):5936-45. doi: 10.1158/0008-5472.CAN-08-4914. Epub 2009 Jun 23. Cancer Res. 2009. PMID: 19549897 Free PMC article. - An early biomarker and potential therapeutic target of RUNX 3 hypermethylation in breast cancer, a system review and meta-analysis.
Lu DG, Ma YM, Zhu AJ, Han YW. Lu DG, et al. Oncotarget. 2017 Mar 28;8(13):22166-22174. doi: 10.18632/oncotarget.13125. Oncotarget. 2017. PMID: 27825140 Free PMC article. Review. - Human breast epithelial stem cells and their regulation.
Kalirai H, Clarke RB. Kalirai H, et al. J Pathol. 2006 Jan;208(1):7-16. doi: 10.1002/path.1881. J Pathol. 2006. PMID: 16294373 Review.
Cited by
- Genetic differences in transcript responses to low-dose ionizing radiation identify tissue functions associated with breast cancer susceptibility.
Snijders AM, Marchetti F, Bhatnagar S, Duru N, Han J, Hu Z, Mao JH, Gray JW, Wyrobek AJ. Snijders AM, et al. PLoS One. 2012;7(10):e45394. doi: 10.1371/journal.pone.0045394. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077491 Free PMC article. - High-fat or ethinyl-oestradiol intake during pregnancy increases mammary cancer risk in several generations of offspring.
de Assis S, Warri A, Cruz MI, Laja O, Tian Y, Zhang B, Wang Y, Huang TH, Hilakivi-Clarke L. de Assis S, et al. Nat Commun. 2012;3:1053. doi: 10.1038/ncomms2058. Nat Commun. 2012. PMID: 22968699 Free PMC article. - TET repression and increased DNMT activity synergistically induce aberrant DNA methylation.
Takeshima H, Niwa T, Yamashita S, Takamura-Enya T, Iida N, Wakabayashi M, Nanjo S, Abe M, Sugiyama T, Kim YJ, Ushijima T. Takeshima H, et al. J Clin Invest. 2020 Oct 1;130(10):5370-5379. doi: 10.1172/JCI124070. J Clin Invest. 2020. PMID: 32663196 Free PMC article. - Epigenetics in Inflammatory Breast Cancer: Biological Features and Therapeutic Perspectives.
Faldoni FLC, Rainho CA, Rogatto SR. Faldoni FLC, et al. Cells. 2020 May 8;9(5):1164. doi: 10.3390/cells9051164. Cells. 2020. PMID: 32397183 Free PMC article. Review. - Actions of endocrine-disrupting chemicals on stem/progenitor cells during development and disease.
Kopras E, Potluri V, Bermudez ML, Williams K, Belcher S, Kasper S. Kopras E, et al. Endocr Relat Cancer. 2014 Mar 12;21(2):T1-12. doi: 10.1530/ERC-13-0360. Print 2014 Apr. Endocr Relat Cancer. 2014. PMID: 24280134 Free PMC article. Review.
References
- Fenton SE. Endocrine-disrupting compounds and mammary gland development: early exposure and later life consequences. Endocrinology. 2006;147:S18–S24. - PubMed
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. - PubMed
- Newbold RR, Padilla-Banks E, Jefferson WN. Adverse effects of the model environmental estrogen diethylstilbestrol are transmitted to subsequent generations. Endocrinology. 2006;147:S11–S17. - PubMed
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA098522/CA/NCI NIH HHS/United States
- R01 CA069065/CA/NCI NIH HHS/United States
- P30 CA16058/CA/NCI NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- U01 ES015986/ES/NIEHS NIH HHS/United States
- U54 CA11300/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases