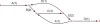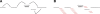ALLPATHS: de novo assembly of whole-genome shotgun microreads - PubMed (original) (raw)
ALLPATHS: de novo assembly of whole-genome shotgun microreads
Jonathan Butler et al. Genome Res. 2008 May.
Abstract
New DNA sequencing technologies deliver data at dramatically lower costs but demand new analytical methods to take full advantage of the very short reads that they produce. We provide an initial, theoretical solution to the challenge of de novo assembly from whole-genome shotgun "microreads." For 11 genomes of sizes up to 39 Mb, we generated high-quality assemblies from 80x coverage by paired 30-base simulated reads modeled after real Illumina-Solexa reads. The bacterial genomes of Campylobacter jejuni and Escherichia coli assemble optimally, yielding single perfect contigs, and larger genomes yield assemblies that are highly connected and accurate. Assemblies are presented in a graph form that retains intrinsic ambiguities such as those arising from polymorphism, thereby providing information that has been absent from previous genome assemblies. For both C. jejuni and E. coli, this assembly graph is a single edge encompassing the entire genome. Larger genomes produce more complicated graphs, but the vast majority of the bases in their assemblies are present in long edges that are nearly always perfect. We describe a general method for genome assembly that can be applied to all types of DNA sequence data, not only short read data, but also conventional sequence reads.
Figures
Figure 1.
Unipath graph of the 1.8-Mb genome of C. jejuni for K = 6000, which is also the best possible assembly of its genome from unpaired reads of length 6001. The genome was treated as linear to simplify computation. Each unipath is labeled with its number of copies (multiplicity) in the genome and with a letter to facilitate discussion. Formally, the graph also includes a reversed copy corresponding to the reverse-complemented sequence (data not shown). The middle horizontal edge represents a 6.2-kb perfect repeat present three times in the genome. This edge is present exactly because the reads are shorter than the repeat. If the reads were longer than 6.2 kb, then the graph would be a single edge. This graph (along with edge sequences and multiplicities) represents exactly what can be known from the data: There are exactly two ways to traverse the graph from end to end, ABCDBCEFCEG and ABCEFCDBCEG, but it is not possible to know from the data which of these alternatives represents the true genome. When K = 20, one instead has 4161 unipaths in total, and the graph is far too tangled to display, or even to separate the genome from its complemented copy.
Figure 2.
Localization. (A) Lines represent unipaths, and curves represent paired-read links between them; from seed, iteratively link to low-copy-number unipaths within a 10-kb radius of it. (B) Reads aligning to these unipaths have partners (red) that dangle in repetitive gaps between them.
Figure 3.
Finding pairs in secondary read cloud. An arbitrary short-fragment read pair is shown (red). If both its reads can be separately subsumed as perfect matches to contigs built from reads from the primary cloud (black), the pair is placed in the secondary read cloud. The black contigs represent all possible ways of combining reads from the primary read cloud using perfect overlaps that are at least K bases long.
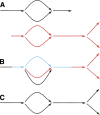
Figure 4.
Merger of sequence graphs in ALLPATHS. The process is iterative. It starts with a collection of sequence graphs, and progressively glues them together. Note the simplest case: all the sequence graphs might consist of single edges. Example: (A) two sequence graphs match at graph and sequence level along common portion consisting of bubble extended on both ends; (B) the algorithm identifies a common linear stretch (blue) that extends from a source on one graph to a sink on the other, then glues the graphs along this stretch; however, parallel black and red edges at the bottom are not yet glued; (C) now these edges are zipped up.
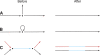
Figure 5.
Editing assembly graphs. Assembly graphs are edited to improve their quality. (A) Clean-up operations, for example, removal of short “hanging ends,” like the middle vertical edge; other clean-up operations include deletion of sequence that is not covered by paired reads and deletion of tiny graph components. (B) Disambiguation operations. Here, given sufficient paired-read links from the left to the right edge, the precise number of copies of the loop edge may be determined, and it may then be unrolled, thereby replacing all three edges by a single edge. (C) Pulling-apart operations. If paired-read links go from the left red edge to the right red edge, and from the left black edge to the right black edge, but not from red to black or black to red, the middle edge may be duplicated, yielding as output two composite edges.
Figure 6.
ALLPATHS assemblies of simulated 30-base microreads. Four assembly graphs or parts are shown, with edges color-coded to reflect length: (gray) <100; (black) 100–1000; (red) 1000–10,000; (magenta) >10,000. (Image created using Graphviz [Low 2004].) None of the graph parts have errors, but some have ambiguities. (A) Assembly of E. coli is one edge. (B) 1.1-Mb component from assembly of 21-Mb fungal genome Y. lipolytica. Five ambiguities are seen: All are loops labeled “1” corresponding to mononucleotide runs whose exact length is unknown. (C) Tiny section of component of assembly of diploid human 10-Mb region. Five ambiguities are seen: All are bubbles arising from SNPs, which are intrinsic to the genome. (D) Correct but tangled component of Pichia stipitis assembly. There is a unique path between the two light blue vertices that matches the reference perfectly.
Figure 7.
Unipaths in a genome. A hypothetical genome has 12 _K_-mers, represented here as vertices. There are five unipaths, one for each of five colors. Vertex L has indegree 2 and vertex R has outdegree 2, delineating branches.
Similar articles
- Velvet: algorithms for de novo short read assembly using de Bruijn graphs.
Zerbino DR, Birney E. Zerbino DR, et al. Genome Res. 2008 May;18(5):821-9. doi: 10.1101/gr.074492.107. Epub 2008 Mar 18. Genome Res. 2008. PMID: 18349386 Free PMC article. - Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads.
Wick RR, Judd LM, Gorrie CL, Holt KE. Wick RR, et al. PLoS Comput Biol. 2017 Jun 8;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. eCollection 2017 Jun. PLoS Comput Biol. 2017. PMID: 28594827 Free PMC article. - Benchmarking of de novo assembly algorithms for Nanopore data reveals optimal performance of OLC approaches.
Cherukuri Y, Janga SC. Cherukuri Y, et al. BMC Genomics. 2016 Aug 22;17 Suppl 7(Suppl 7):507. doi: 10.1186/s12864-016-2895-8. BMC Genomics. 2016. PMID: 27556636 Free PMC article. - De novo assembly of short sequence reads.
Paszkiewicz K, Studholme DJ. Paszkiewicz K, et al. Brief Bioinform. 2010 Sep;11(5):457-72. doi: 10.1093/bib/bbq020. Epub 2010 Aug 19. Brief Bioinform. 2010. PMID: 20724458 Review. - State of the art de novo assembly of human genomes from massively parallel sequencing data.
Li Y, Hu Y, Bolund L, Wang J. Li Y, et al. Hum Genomics. 2010 Apr;4(4):271-7. doi: 10.1186/1479-7364-4-4-271. Hum Genomics. 2010. PMID: 20511140 Free PMC article. Review.
Cited by
- Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi.
Cacho RA, Tang Y, Chooi YH. Cacho RA, et al. Front Microbiol. 2015 Jan 14;5:774. doi: 10.3389/fmicb.2014.00774. eCollection 2014. Front Microbiol. 2015. PMID: 25642215 Free PMC article. Review. - A hybrid approach for the automated finishing of bacterial genomes.
Bashir A, Klammer A, Robins WP, Chin CS, Webster D, Paxinos E, Hsu D, Ashby M, Wang S, Peluso P, Sebra R, Sorenson J, Bullard J, Yen J, Valdovino M, Mollova E, Luong K, Lin S, LaMay B, Joshi A, Rowe L, Frace M, Tarr CL, Turnsek M, Davis BM, Kasarskis A, Mekalanos JJ, Waldor MK, Schadt EE. Bashir A, et al. Nat Biotechnol. 2012 Jul 1;30(7):701-707. doi: 10.1038/nbt.2288. Nat Biotechnol. 2012. PMID: 22750883 Free PMC article. - The genome of Diuraphis noxia, a global aphid pest of small grains.
Nicholson SJ, Nickerson ML, Dean M, Song Y, Hoyt PR, Rhee H, Kim C, Puterka GJ. Nicholson SJ, et al. BMC Genomics. 2015 Jun 5;16:429. doi: 10.1186/s12864-015-1525-1. BMC Genomics. 2015. PMID: 26044338 Free PMC article. - Revealing Missing Human Protein Isoforms Based on Ab Initio Prediction, RNA-seq and Proteomics.
Hu Z, Scott HS, Qin G, Zheng G, Chu X, Xie L, Adelson DL, Oftedal BE, Venugopal P, Babic M, Hahn CN, Zhang B, Wang X, Li N, Wei C. Hu Z, et al. Sci Rep. 2015 Jul 9;5:10940. doi: 10.1038/srep10940. Sci Rep. 2015. PMID: 26156868 Free PMC article. - Finished Genome Sequence of Bacillus cereus Strain 03BB87, a Clinical Isolate with B. anthracis Virulence Genes.
Johnson SL, Minogue TD, Teshima H, Davenport KW, Shea AA, Miner HL, Wolcott MJ, Chain PS. Johnson SL, et al. Genome Announc. 2015 Jan 15;3(1):e01446-14. doi: 10.1128/genomeA.01446-14. Genome Announc. 2015. PMID: 25593267 Free PMC article.
References
- Batzoglou S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Berger B., Mesirov J.P., Lander E.S., Mesirov J.P., Lander E.S., Lander E.S. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12:177–189. - PMC - PubMed
- Jeck W.R., Reinhardt J.A., Baltrus D.A., Hickenbotham M.T., Magrini V., Mardis E.R., Dangl J.L., Jones C.D., Reinhardt J.A., Baltrus D.A., Hickenbotham M.T., Magrini V., Mardis E.R., Dangl J.L., Jones C.D., Baltrus D.A., Hickenbotham M.T., Magrini V., Mardis E.R., Dangl J.L., Jones C.D., Hickenbotham M.T., Magrini V., Mardis E.R., Dangl J.L., Jones C.D., Magrini V., Mardis E.R., Dangl J.L., Jones C.D., Mardis E.R., Dangl J.L., Jones C.D., Dangl J.L., Jones C.D., Jones C.D. Extending assembly of short DNA sequences to handle error. Bioinformatics. 2007;23:2942–2944. - PubMed
- Johnson D.S., Mortazavi A., Myers R.M., Wold B., Mortazavi A., Myers R.M., Wold B., Myers R.M., Wold B., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. - PubMed
- Low G. 2004. Graphviz. http://www.graphviz.org.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous