The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila - PubMed (original) (raw)
The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila
Sarah K Bowman et al. Dev Cell. 2008 Apr.
Abstract
In both vertebrates and insects, neurons typically arise from neural stem cells or terminally dividing intermediate progenitors. Here, we describe another mode of neurogenesis where neural stem cells generate secondary precursors that undergo multiple rounds of self-renewing transit-amplifying divisions. We identify the Posterior Asense-Negative (PAN) neuroblasts, which do not express the transcription factors Asense or Prospero. PAN neuroblasts rely on the segregating determinants Numb and Brat to generate smaller, secondary neuroblasts that in turn give rise to ganglion mother cells (GMCs) and neurons throughout larval development. In brat or numb mutants, misspecified secondary neuroblasts are unable to produce differentiated progeny and initiate tumor-like overgrowth. In prospero mutants, however, tumors arise from GMCs while secondary neuroblasts are correctly specified. Our data describe a transit-amplifying lineage in the Drosophila nervous system and suggest that different vulnerabilities in intermediate cell types can affect the outcome of tumor suppressor loss in stem cell lineages.
Figures
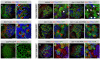
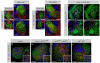
Figure 1. brat Overgrowth Originates in PAN Neuroblast Lineages
Immunostainings of third-instar brains labeled with indicated markers (gray boxes). For each genotype, an overview of the brain lobe and a detail of neuroblast lineages are shown. (A and B) Wild-type brains contain PAN neuroblasts with long lineages of Ase+ progeny ([A] and [A′], outline); ectopic expression of Ase eliminates both ([B] and [B′], outline). (C and C′) A brat zygotic mutant brain is overgrown with PAN neuroblasts. (D–G) Brains with GFP reporting _insc_-Gal4 expression. Control brains are well organized and contain PAN neuroblasts ([D] and [D′], arrowhead). Brat knockdown results in disorganized overgrowth with ectopic PAN neuroblasts ([E] and [E′], arrowheads). Expressing Ase in all neuroblasts eliminates PAN neuroblasts (F and F′). Ectopic expression of Ase prevents the overgrowth normally caused by Brat knockdown (G and G′). (H and I) Brains with GFP reporting _ase_-Gal4 expression. Neither control brains (H and H′) nor Brat knockdown brains (I and I′) show disorganized overgrowth phenotypes. Scale bars: (A)–(I), 50 μm; (A′)–(I′), 10 μm.
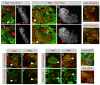
Figure 2. PAN Lineages Produce Intermediate Progenitors
Immunostainings of MARCM clones in central brain neuroblasts labeled with indicated markers (gray boxes). p-H3: phospho-histone H3. Neuroblast clones are 3D structures, so two separate optical sections from a Z-stack through each clone are shown. The most superficial section is labeled 0 μm. (A–D) Neuroblast clones are reported by GFP expression. In 24 hr clones (A) and 48 hr clones (B), Ase+ neuroblasts produce Ase+ and Elav+ progeny. A single Ase+ cell divides symmetrically in size and adjacent to the neuroblast ([B′], arrowhead). In 24 hr PAN neuroblast clones, the neuroblast produces Ase− progeny and Ase+ progeny, but almost no Elav+ progeny (C). Forty-eight hour PAN neuroblast clones contain a small number of Ase− cells, and large numbers of Ase+ and Elav+ cells (D). All mitotic cells are Ase+, several are far away from the neuroblast, and one divides asymmetrically in size ([D′], arrowhead). Scale bars: 10 μm.
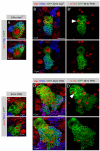
Figure 3. PAN Lineages Contain Secondary Neuroblasts
Immunostainings of larval brains labeled with indicated markers (gray boxes). Primary neuroblasts are marked with a star. (A and B) Forty-eight hour MARCM clones in central brain neuroblasts. Two separate optical sections from a Z-stack through each clone are shown. Neuroblast clones are reported by GFP expression. (A and A′) Ase+ neuroblasts exclude Pros from the nucleus, and generate Ase+Pros+ GMCs (open arrowhead) and Ase−Pros+ neurons (closed arrowhead). (B and B′) PAN neuroblasts do not have nuclear Pros. All Ase− (open arrowhead) and many Ase+ (closed arrowhead) daughters do not have nuclear Pros. GMCs are Ase+Pros+ (closed arrow) and neurons are Ase−Pros+ (open arrow). (C and D) Many PAN neuroblast progeny express the neuroblast markers CycE (C) and Dpn (D). (E–H) Neuroblast daughter cells in mitosis (arrowheads). Mira is not present in mitotic GMCs (E′) but segregates asymmetrically in some progeny of the PAN neuroblast (F′). Similarly, Pros is cytoplasmic in mitotic GMCs (G′) but asymmetric cortical in some PAN progeny (H′). (I and J) Twenty-four hour (I) and forty-eight hour (J) secondary neuroblast clones contain multiple Ase+ cells (I) and multiple neurons (J). Scale bars: 10 μm.
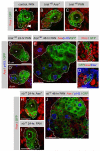
Figure 4. Immature Ase− Secondary Neuroblasts Enter Mitosis in brat
Immunostainings of MARCM clones reported by GFP expression and labeled with indicated markers (gray boxes). Stars indicate primary neuroblasts and circles indicate daughter cells. (A–C) Pros in 24 hr clones. In wild-type PAN neuroblast clones, Pros is nuclear in some daughters ([A], arrowhead). In brat Ase+ clones, Pros is nuclear in all daughters (B), but in brat PAN neuroblast clones, no daughters have nuclear Pros (C). (D and E) Mitotic Ase− cells in brat PAN neuroblast clones. In 24 hr clones, Ase− secondary neuroblasts enter mitosis (D′). In 48 hr clones, mitotic Ase− cells increase in size and number (E). (F and G) brat secondary neuroblast clones. Differentiated Pros+ cells (F) and Ase+ and Elav+ cells (G) are created in the absence of brat. (H–J) numb clones. In 24 hr numb Ase+ clones, all daughters have nuclear Pros (H). Pros is not nuclear in daughters of numb PAN neuroblasts (I). Ase− daughters enter mitosis in numb 48 hr PAN neuroblast clones. Scale bars: 10 μm.
Figure 5. Pros Is Not Required for Maturation of Secondary Neuroblasts
Immunostainings labeled with indicated markers (gray boxes). (A–D) pros MARCM clones reported by GFP expression. Stars indicate primary neuroblasts. Two separate optical sections from a Z-stack through each clone are shown. Loss of Pros in Ase+ lineages results in failure to generate neurons (A and B). In PAN lineages, Pros is not required to generate Ase+ daughters, but neurons fail to differentiate (C and D). (E and F) _pros_-Gal4 expression reported by GFP. _pros_-Gal4 is not expressed in the PAN neuroblasts (E) or their Ase− daughters (circles, [F]). (G–J) PAN lineage behavior in other tumor models from early third-instar larval brains. Insets: Ase+ neuroblasts of the ventral nerve cord. Expression of membrane-targeted aPKC (H) or loss of AurA (I) results in overgrowth in both Ase+ and PAN lineages, but loss of Lgl affects PAN lineages more strongly (J). Scale bars: (A)–(D) and (F), 10 μm; (E) and (G)–(J), 50 μm.
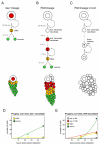
Figure 6. Models of Neuroblast Lineages in the Central Brain
(A–C) Lineage diagrams with estimated time for each stage and cartoons of corresponding neuroblast clones. (A) Self-renewing Ase+ neuroblasts produce progeny that follow the standard GMC-neuron progression. (B) PAN neuroblasts generate Ase− secondary neuroblasts that mature into self-renewing Ase+ secondary neuroblasts. These transit-amplifying cells produce GMCs, which divide terminally to produce two neurons. (C) In the absence of Brat, Ase− secondary neuroblasts are unable to become Ase+. Upon mitotic entry, they become tumor-initiating cells. (D and E) Plots showing production of neuroblast progeny over time. Production of neurons in Ase+ lineages is linear (D), while production of neurons in PAN lineages is exponential (E). Plots were created using Matlab and computer-modeled neuroblast lineages (see Supplemental Experimental Procedures for details). Colored circles correspond to the cell numbers recorded in Table 2.
Similar articles
- Protein phosphatase 2A regulates self-renewal of Drosophila neural stem cells.
Wang C, Chang KC, Somers G, Virshup D, Ang BT, Tang C, Yu F, Wang H. Wang C, et al. Development. 2009 Jul;136(13):2287-96. doi: 10.1242/dev.035758. Development. 2009. PMID: 19502489 - Distinct functions of human numb isoforms revealed by misexpression in the neural stem cell lineage in the Drosophila larval brain.
Toriya M, Tokunaga A, Sawamoto K, Nakao K, Okano H. Toriya M, et al. Dev Neurosci. 2006;28(1-2):142-55. doi: 10.1159/000090760. Dev Neurosci. 2006. PMID: 16508311 - Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells.
Boone JQ, Doe CQ. Boone JQ, et al. Dev Neurobiol. 2008 Aug;68(9):1185-95. doi: 10.1002/dneu.20648. Dev Neurobiol. 2008. PMID: 18548484 Free PMC article. - Diversifying neural cells through order of birth and asymmetry of division.
Zhong W. Zhong W. Neuron. 2003 Jan 9;37(1):11-4. doi: 10.1016/s0896-6273(02)01178-9. Neuron. 2003. PMID: 12526768 Review. - Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development.
Reichert H. Reichert H. Results Probl Cell Differ. 2011;53:529-46. doi: 10.1007/978-3-642-19065-0_21. Results Probl Cell Differ. 2011. PMID: 21630158 Review.
Cited by
- Light Stimuli and Circadian Clock Affect Neural Development in Drosophila melanogaster.
Dapergola E, Menegazzi P, Raabe T, Hovhanyan A. Dapergola E, et al. Front Cell Dev Biol. 2021 Mar 5;9:595754. doi: 10.3389/fcell.2021.595754. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33763414 Free PMC article. - dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila.
Weng M, Golden KL, Lee CY. Weng M, et al. Dev Cell. 2010 Jan 19;18(1):126-35. doi: 10.1016/j.devcel.2009.12.007. Dev Cell. 2010. PMID: 20152183 Free PMC article. - The Ets protein Pointed P1 represses Asense expression in type II neuroblasts by activating Tailless.
Chen R, Deng X, Zhu S. Chen R, et al. PLoS Genet. 2022 Jan 31;18(1):e1009928. doi: 10.1371/journal.pgen.1009928. eCollection 2022 Jan. PLoS Genet. 2022. PMID: 35100262 Free PMC article. - Illumination of neural development by in vivo clonal analysis.
Xu M, Wang J, Guo X, Li T, Kuang X, Wu QF. Xu M, et al. Cell Regen. 2018 Oct 12;7(2):33-39. doi: 10.1016/j.cr.2018.09.001. eCollection 2018 Dec. Cell Regen. 2018. PMID: 30671228 Free PMC article. Review. - Faster, higher, stronger: timely and robust cell fate/identity commitment in stem cell lineages.
Liu K, Xu K, Song Y. Liu K, et al. Open Biol. 2019 Feb 28;9(2):180243. doi: 10.1098/rsob.180243. Open Biol. 2019. PMID: 30958098 Free PMC article. Review.
References
- Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. - PubMed
- Badenhorst P, Finch JT, Travers AA. Tramtrack co-operates to prevent inappropriate neural development in Drosophila. Mech. Dev. 2002;117:87–101. - PubMed
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. - PubMed
- Berdnik D, Knoblich JA. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 2002;12:640–647. - PubMed
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell. 2002;3:221–231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases