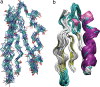Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR - PubMed (original) (raw)
Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR
W Trent Franks et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Magic-angle spinning (MAS) solid-state NMR (SSNMR) techniques have emerged in recent years for solving complete structures of uniformly labeled proteins lacking macroscopic order. Strategies used thus far have relied primarily on semiquantitative distance restraints, analogous to the nuclear Overhauser effect (NOE) routinely used in solution NMR. Here, we present a complementary approach for using relative orientations of molecular fragments, determined from dipolar line shapes. Whereas SSNMR distance restraints typically have an uncertainty of approximately 1 A, the tensor-based experiments report on relative vector (pseudobond) angles with precision of a few degrees. By using 3D techniques of this type, vector angle (VEAN) restraints were determined for the majority of the 56-residue B1 immunoglobulin binding domain of protein G [protein GB1 (a total of 47 HN-HN, 49 HN-HC, and 12 HA-HB restraints)]. By using distance restraints alone in the structure calculations, the overall backbone root-mean-square deviation (bbRMSD) was 1.01 +/- 0.13 A (1.52 +/- 0.12 A for all heavy atoms), which improved to 0.49 +/- 0.05 A (1.19 +/- 0.07 A) on the addition of empirical chemical shift [torsion angle likelihood obtained from shift and sequence similarity (TALOS)] restraints. VEAN restraints further improved the ensemble to 0.31 +/- 0.06 A bbRMSD (1.06 +/- 0.07 A); relative to the structure with distances alone, most of the improvement remained (bbRMSD 0.64 +/- 0.09 A; 1.29 +/- 0.07 A) when TALOS restraints were removed before refinement. These results represent significant progress toward atomic-resolution protein structure determination by SSNMR, capabilities that can be applied to a large range of membrane proteins and fibrils, which are often not amenable to solution NMR or x-ray crystallography.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
2D 13C–13C correlation spectra of sparsely 13C-labeled samples of GB1, 750 MHz 1H frequency, 300 ms longitudinal DARR mixing, 12.5 kHz MAS rate. (a) Aliphatic region, sample E. (b) Expansion of the near-diagonal CA region (sample E), illustrating medium and long-range CA[_i_]-CA[_j_] correlations. (c) Aliphatic region, sample D. (d) Expansion of the methyl region (sample D). Detailed acquisition and processing parameters are described in
SI Figs. 7–9
. Peak intensities are interpreted semiempirically in terms of internuclear distances.
Fig. 2.
Ensemble of the 10 lowest energy structures of GB1, calculated from a total of 7,826 13C–13C, 15N–15N, and 1H–1H distance restraints. The bbRMSD for all residues is 1.01 ± 0.13 Å, and the heavy atom RMSD is 1.52 ± 0.12 Å. (a) Line representation including all backbone carbon (cyan), amide nitrogen (blue), amide proton (white) and carbonyl oxygen (red) atoms. (b) Diagram representation indicating ordered secondary structure elements (helix in purple, strands in yellow, turns in cyan, coil in white).
Fig. 3.
Dipolar line shapes from 3D dipolar-shift correlation spectra of GB1, used to derive vector angle (VEAN) restraints. Experimental HN-HACA line shapes (blue) were fit to simulations of the spin dynamics (red), as a function of the relative orientation of the two 1H-X dipole vectors. The best-fit values for the VEANs are θ = 14.0° ± 2.0° for T51 (a); θ = 21.2° ± 3.1° for V39 (b); θ = 19.5° ± 2.1° for T49 (c); and θ = 42.0° ± 6.3° for V29 (d). Standard errors are determined by Monte Carlo analysis.
Fig. 4.
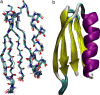
Ensemble of 10 GB1 structures determined from distance, TALOS (chemical shift), and VEAN restraints (bbRMSD 0.31 ± 0.06 Å, heavy atom RMSD 1.06 ± 0.07 Å). (a) Line representation; (b) diagram representation. Color coding is identical to Fig. 2.
Fig. 5.
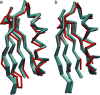
Structural alignment of high-resolution SSNMR ensemble (cyan) and the trigonal form crystal structure (2QMT; red). (a) With all residues aligned, the bbRMSD is 1.4 Å. (b) Alignment excluding residues 1, 9–14, and 39–41 (1.1 Å bbRMSD), demonstrating that residues in the β1-β2 turn and helix-β3 loop disrupt the relative positioning of helix and four-stranded β-sheet.
Similar articles
- Exploring the limits of precision and accuracy of protein structures determined by nuclear magnetic resonance spectroscopy.
Clore GM, Robien MA, Gronenborn AM. Clore GM, et al. J Mol Biol. 1993 May 5;231(1):82-102. doi: 10.1006/jmbi.1993.1259. J Mol Biol. 1993. PMID: 8496968 - Magic angle spinning NMR structure determination of proteins from pseudocontact shifts.
Li J, Pilla KB, Li Q, Zhang Z, Su X, Huber T, Yang J. Li J, et al. J Am Chem Soc. 2013 Jun 5;135(22):8294-303. doi: 10.1021/ja4021149. Epub 2013 May 24. J Am Chem Soc. 2013. PMID: 23646876 - Backbone conformational constraints in a microcrystalline U-15N-labeled protein by 3D dipolar-shift solid-state NMR spectroscopy.
Franks WT, Wylie BJ, Stellfox SA, Rienstra CM. Franks WT, et al. J Am Chem Soc. 2006 Mar 15;128(10):3154-5. doi: 10.1021/ja058292x. J Am Chem Soc. 2006. PMID: 16522090 - Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.
Zhang R, Mroue KH, Ramamoorthy A. Zhang R, et al. Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review. - Dihedral Angle Measurements for Structure Determination by Biomolecular Solid-State NMR Spectroscopy.
van der Wel PCA. van der Wel PCA. Front Mol Biosci. 2021 Dec 6;8:791090. doi: 10.3389/fmolb.2021.791090. eCollection 2021. Front Mol Biosci. 2021. PMID: 34938776 Free PMC article. Review.
Cited by
- β-Helical architecture of cytoskeletal bactofilin filaments revealed by solid-state NMR.
Vasa S, Lin L, Shi C, Habenstein B, Riedel D, Kühn J, Thanbichler M, Lange A. Vasa S, et al. Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):E127-36. doi: 10.1073/pnas.1418450112. Epub 2014 Dec 30. Proc Natl Acad Sci U S A. 2015. PMID: 25550503 Free PMC article. - Zero-quantum stochastic dipolar recoupling in solid state nuclear magnetic resonance.
Qiang W, Tycko R. Qiang W, et al. J Chem Phys. 2012 Sep 14;137(10):104201. doi: 10.1063/1.4749258. J Chem Phys. 2012. PMID: 22979851 Free PMC article. - Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy.
Brown LS, Ladizhansky V. Brown LS, et al. Protein Sci. 2015 Sep;24(9):1333-46. doi: 10.1002/pro.2700. Epub 2015 May 27. Protein Sci. 2015. PMID: 25973959 Free PMC article. Review. - Ubiquitin immobilized on mesoporous MCM41 silica surfaces - Analysis by solid-state NMR with biophysical and surface characterization.
Adiram-Filiba N, Schremer A, Ohaion E, Nadav-Tsubery M, Lublin-Tennenbaum T, Keinan-Adamsky K, Goobes G. Adiram-Filiba N, et al. Biointerphases. 2017 May 31;12(2):02D414. doi: 10.1116/1.4983273. Biointerphases. 2017. PMID: 28565916 Free PMC article. - Backbone assignment of perdeuterated proteins using long-range H/C-dipolar transfers.
Linser R. Linser R. J Biomol NMR. 2012 Feb;52(2):151-8. doi: 10.1007/s10858-011-9593-2. Epub 2011 Dec 14. J Biomol NMR. 2012. PMID: 22167467
References
- McDermott A, et al. Partial NMR assignments for uniformly-13C,15N-enriched BPTI in the solid state. J Biomol NMR. 2000;16:209–219. - PubMed
- Castellani F, et al. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. - PubMed
- Lange A, et al. A concept for rapid protein-structure determination by solid-state NMR spectroscopy. Angew Chem Int Ed. 2005;44:2089–2092. - PubMed
- Zech SG, Wand AJ, McDermott AE. Protein structure determination by high-resolution solid-state NMR spectroscopy: Application to microcrystalline ubiquitin. J Am Chem Soc. 2005;127:8618–8626. - PubMed
- Etzkorn M, et al. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes studied by solid-state nmr. Angew Chem Int Ed. 2007;46:459–462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources