RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members - PubMed (original) (raw)
RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members
Rays H Y Jiang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Pathogens secrete effector molecules that facilitate the infection of their hosts. A number of effectors identified in plant pathogenic Phytophthora species possess N-terminal motifs (RXLR-dEER) required for targeting these effectors into host cells. Here, we bioinformatically identify >370 candidate effector genes in each of the genomes of P. sojae and P. ramorum. A single superfamily, termed avirulence homolog (Avh) genes, accounts for most of the effectors. The Avh proteins show extensive sequence divergence but are all related and likely evolved from a common ancestor by rapid duplication and divergence. More than half of the Avh proteins contain conserved C-terminal motifs (termed W, Y, and L) that are usually arranged as a module that can be repeated up to eight times. The Avh genes belong to the most rapidly evolving part of the genome, and they are nearly always located at synteny breakpoints. The superfamily includes all experimentally identified oomycete effector and avirulence genes, and its rapid pace of evolution is consistent with a role for Avh proteins in interaction with plant hosts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
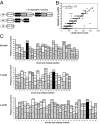
Relationship between Avh proteins. (A) Pairwise similarity of Avh groups (AGs). For every pair of groups, the two most similar Avh proteins were used for the sequence similarity analysis. The AGs from left to right and from top to bottom are AG_1_1 to AG_1_9, AG_2 to AG_78, and AG_83. (B) Pairwise similarity of randomly selected 1,000 P. sojae proteins. (C) Phylogram constructed with several AGs. The P. sojae Avh proteins are shaded in gray. The unrooted phylogram is based on NJ analysis. All major clades (indicated by black dots) are consistent with similarity groupings. Confidence of groupings was estimated by using 1,000 bootstrap replicates; numbers next to the branching point indicate the percentage of replicates supporting each branch. On the branch tip, the individual Avh proteins are indicated.
Fig. 2.
Conserved C-terminal motifs. (A) Motifs in Avh proteins; 36% of the Avh proteins have a combination of W, Y, and L motifs, 22% have only W motifs, and the remaining 42% do not have identifiable W, Y, or L motifs. SP, signal peptide. (B) The correlation between the number of W motifs and the size of the protein. (C) The consensus sequence of W, Y, and L motifs. The size of the histograms indicates the frequency of the occurrence of the amino acid residue. The most abundant four residues are shown if their frequencies are >2%. The most frequent amino acid at each position is shaded if the frequency is >20%. The most abundant W, Y, and L residues are shaded in black.
Fig. 3.
High level of sequence divergence and polymorphisms. (A) Sequence divergence of Avh proteins. Sequence divergence is defined as one sequence identity. PsAvh and PrAvh proteins were compared against P. ramorum and P. sojae proteins, respectively. Each radius ranges from 0 (center) to 80 (outer circle) representing 100–20% (or less) identity. The distribution along each circumference is random. A set of 1,000 randomly selected P. sojae proteins was compared against P. ramorum. (B) Positively selected sites in Avh proteins. Codeml model 2a was used to detect positively selected sites (shaded in black). The motifs are indicated by asterisks underneath. The analysis was performed on alleles of Avr1b (9) and close paralogs of PrAvh302.
Fig. 4.
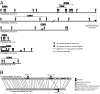
Synteny breakpoints at the locations of Avh genes. (A) Distribution of Avh genes represented as indels. Six scaffolds containing more than two PsAvh genes are shown. PsScaffold_36 and PrScaffold_50 contain the largest clusters of Avh genes in P. sojae and P. ramorum, respectively. Paralogs belonging to the same family are indicated by lines. (B) One deletion and one rearrangement of PsAvh genes in the P. sojae scaffold_23. Orthologs are connected by thin lines.
Similar articles
- Recognition of Phytophthora infestans Avr4 by potato R4 is triggered by C-terminal domains comprising W motifs.
VAN Poppel PM, Jiang RH, Sliwka J, Govers F. VAN Poppel PM, et al. Mol Plant Pathol. 2009 Sep;10(5):611-20. doi: 10.1111/j.1364-3703.2009.00556.x. Mol Plant Pathol. 2009. PMID: 19694952 Free PMC article. - Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a.
Qutob D, Tedman-Jones J, Dong S, Kuflu K, Pham H, Wang Y, Dou D, Kale SD, Arredondo FD, Tyler BM, Gijzen M. Qutob D, et al. PLoS One. 2009;4(4):e5066. doi: 10.1371/journal.pone.0005066. Epub 2009 Apr 3. PLoS One. 2009. PMID: 19343173 Free PMC article. - Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b.
Dou D, Kale SD, Wang X, Chen Y, Wang Q, Wang X, Jiang RH, Arredondo FD, Anderson RG, Thakur PB, McDowell JM, Wang Y, Tyler BM. Dou D, et al. Plant Cell. 2008 Apr;20(4):1118-33. doi: 10.1105/tpc.107.057067. Epub 2008 Apr 4. Plant Cell. 2008. PMID: 18390593 Free PMC article. - Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans.
Birch PR, Armstrong M, Bos J, Boevink P, Gilroy EM, Taylor RM, Wawra S, Pritchard L, Conti L, Ewan R, Whisson SC, van West P, Sadanandom A, Kamoun S. Birch PR, et al. J Exp Bot. 2009;60(4):1133-40. doi: 10.1093/jxb/ern353. Epub 2009 Feb 9. J Exp Bot. 2009. PMID: 19204033 Review. - Phytophthora genomics: the plant destroyers' genome decoded.
Govers F, Gijzen M. Govers F, et al. Mol Plant Microbe Interact. 2006 Dec;19(12):1295-301. doi: 10.1094/MPMI-19-1295. Mol Plant Microbe Interact. 2006. PMID: 17153913 Review.
Cited by
- Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica.
Jiang RH, de Bruijn I, Haas BJ, Belmonte R, Löbach L, Christie J, van den Ackerveken G, Bottin A, Bulone V, Díaz-Moreno SM, Dumas B, Fan L, Gaulin E, Govers F, Grenville-Briggs LJ, Horner NR, Levin JZ, Mammella M, Meijer HJ, Morris P, Nusbaum C, Oome S, Phillips AJ, van Rooyen D, Rzeszutek E, Saraiva M, Secombes CJ, Seidl MF, Snel B, Stassen JH, Sykes S, Tripathy S, van den Berg H, Vega-Arreguin JC, Wawra S, Young SK, Zeng Q, Dieguez-Uribeondo J, Russ C, Tyler BM, van West P. Jiang RH, et al. PLoS Genet. 2013 Jun;9(6):e1003272. doi: 10.1371/journal.pgen.1003272. Epub 2013 Jun 13. PLoS Genet. 2013. PMID: 23785293 Free PMC article. - Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen.
Rafiqi M, Gan PH, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, Hardham AR, Dodds PN. Rafiqi M, et al. Plant Cell. 2010 Jun;22(6):2017-32. doi: 10.1105/tpc.109.072983. Epub 2010 Jun 4. Plant Cell. 2010. PMID: 20525849 Free PMC article. - Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp.).
Joly DL, Feau N, Tanguay P, Hamelin RC. Joly DL, et al. BMC Genomics. 2010 Jul 8;11:422. doi: 10.1186/1471-2164-11-422. BMC Genomics. 2010. PMID: 20615251 Free PMC article. - Draft Genome Sequence for the Tree Pathogen Phytophthora plurivora.
Vetukuri RR, Tripathy S, Malar C M, Panda A, Kushwaha SK, Chawade A, Andreasson E, Grenville-Briggs LJ, Whisson SC. Vetukuri RR, et al. Genome Biol Evol. 2018 Sep 1;10(9):2432-2442. doi: 10.1093/gbe/evy162. Genome Biol Evol. 2018. PMID: 30060094 Free PMC article. - Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif.
Godfrey D, Böhlenius H, Pedersen C, Zhang Z, Emmersen J, Thordal-Christensen H. Godfrey D, et al. BMC Genomics. 2010 May 20;11:317. doi: 10.1186/1471-2164-11-317. BMC Genomics. 2010. PMID: 20487537 Free PMC article.
References
- Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 2002;10:462–469. - PubMed
- Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. - PubMed
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. - PubMed
- Erwin DC, Ribeiro OK. Phytophthora Diseases Worldwide. St. Paul, MN: Am Phytopathol Soc; 1996.
- Rizzo DM, Garbelotto M, Hansen EM. Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu Rev Phytopathol. 2005;43:309–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources