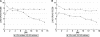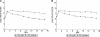Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine - PubMed (original) (raw)
Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine
Shari A Jones et al. Infect Immun. 2008 Jun.
Abstract
Mutant screens and transcriptome studies led us to consider whether the metabolism of glucose polymers, i.e., maltose, maltodextrin, and glycogen, is important for Escherichia coli colonization of the intestine. By using the streptomycin-treated mouse model, we found that catabolism of the disaccharide maltose provides a competitive advantage in vivo to pathogenic E. coli O157:H7 and commensal E. coli K-12, whereas degradation of exogenous forms of the more complex glucose polymer, maltodextrin, does not. The endogenous glucose polymer, glycogen, appears to play an important role in colonization, since mutants that are unable to synthesize or degrade glycogen have significant colonization defects. In support of the hypothesis that E. coli relies on internal carbon stores to maintain colonization during periods of famine, we found that by providing a constant supply of a readily metabolized sugar, i.e., gluconate, in the animal's drinking water, the competitive disadvantage of E. coli glycogen metabolism mutants is rescued. The results suggest that glycogen storage may be widespread in enteric bacteria because it is necessary for maintaining rapid growth in the intestine, where there is intense competition for resources and occasional famine. An important implication of this study is that the sugars used by E. coli are present in limited quantities in the intestine, making endogenous carbon stores valuable. Thus, there may be merit to combating enteric infections by using probiotics or prebiotics to manipulate the intestinal microbiota in such a way as to limit the availability of sugars preferred by E. coli O157:H7 and perhaps other pathogens.
Figures
FIG. 1.
E. coli EDL933 ppsA pckA malP::mini-Tn_5_-Km2 STM is outcompeted by E. coli EDL933 ppsA pckA during colonization. Strains were fed to mice at 105 CFU each.
FIG. 2.
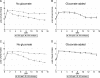
Phenotypes of E. coli EDL933 mutants used in the present study. Growth on maltose or maltodextrin as the sole carbon source was determined in MOPS medium containing 0.2% carbohydrate. Maltose and maltodextrin sensitivity was determined in LB containing 0.2% carbohydrate. The final OD600 values of cultures at 24 h are shown. Oxidation of maltose was determined on Biolog GN2 phenotype plates, as shown in graphlets of relative amounts of substrate oxidized (y axis) versus time. Glycogen accumulation on LB agar containing 2% glucose was visualized by staining with iodine vapor.
FIG. 3.
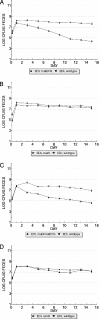
Maltose/maltodextrin transport mutants display colonization defects. (A) E. coli EDL933 Δ(malE-malG) has a colonization defect in competition with E. coli EDL933 wild-type. (B) E. coli EDL933 Δ_malX_ does not exhibit a colonization defect. (C) E. coli EDL933 Δ_malX_ Δ(malE-malG) has a colonization defect in competition with E. coli EDL933 wild-type. (D) E. coli EDL933 Δ_lamB_ does not exhibit a colonization defect. Strains were fed to mice at 105 CFU each.
FIG. 4.
Catabolism of maltose but not maltodextrins is important for competitive colonization of the mouse intestine. (A) E. coli EDL933 Δ(malP-malQ) has colonization defect in competition with E. coli EDL933 wild-type. (B) E. coli EDL933 Δ_malQ_ has colonization defect in competition with E. coli EDL933 wild-type. (C) E. coli EDL933 Δ_malP_ does not exhibit a significant colonization defect. Strains were fed to mice at 105 CFU each.
FIG. 5.
Catabolism of maltose is necessary for E. coli EDL933 to compete efficiently with wild-type E. coli MG1655 in the mouse intestine. (A) E. coli EDL933 wild-type (fed to mice at 105 CFU) initially out competes higher numbers of E. coli MG1655 wild-type (fed at 1010 CFU). (B) E. coli EDL933 Δ_malQ_ (fed at 105 CFU) cannot compete with higher numbers of E. coli MG1655 wild-type (fed at 1010 CFU).
FIG. 6.
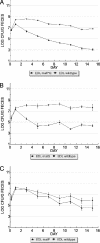
Glycogen synthesis mutants do not compete effectively in the mouse intestine. (A) E. coli EDL933 Δ_glgA_ exhibits colonization defect in competition with E. coli EDL933 wild-type. (B) E. coli EDL933 Δ_glgS_ exhibits colonization defect in competition with E. coli EDL933 wild-type. Strains were fed to mice at 105 CFU each.
FIG. 7.
Colonization defects of glycogen degradation mutants are rescued by adding 2% gluconate to drinking water of mice. (A) E. coli EDL933 Δ_glgP_ and (C) E. coli MG1655 Δ_glgP_ exhibit colonization defects in competition with their respective wild-type parents. (B and D) These colonization defects are rescued by addition of gluconate to the drinking water . Strains were fed to mice at 105 CFU each.
Similar articles
- Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine.
Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Maltby R, et al. PLoS One. 2013;8(1):e53957. doi: 10.1371/journal.pone.0053957. Epub 2013 Jan 17. PLoS One. 2013. PMID: 23349773 Free PMC article. - Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content.
Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, Cohen PS, Conway T, Forano E, Martin C. Bertin Y, et al. Environ Microbiol. 2013 Feb;15(2):610-22. doi: 10.1111/1462-2920.12019. Epub 2012 Nov 6. Environ Microbiol. 2013. PMID: 23126484 Free PMC article. - Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine.
Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. Fabich AJ, et al. Infect Immun. 2008 Mar;76(3):1143-52. doi: 10.1128/IAI.01386-07. Epub 2008 Jan 7. Infect Immun. 2008. PMID: 18180286 Free PMC article. - Fimbriation and curliation in Escherichia coli O157:H7: a paradigm of intestinal and environmental colonization.
Lloyd SJ, Ritchie JM, Torres AG. Lloyd SJ, et al. Gut Microbes. 2012 May-Jun;3(3):272-6. doi: 10.4161/gmic.20661. Epub 2012 May 1. Gut Microbes. 2012. PMID: 22614704 Free PMC article. Review. - A brief overview of Escherichia coli O157:H7 and its plasmid O157.
Lim JY, Yoon J, Hovde CJ. Lim JY, et al. J Microbiol Biotechnol. 2010 Jan;20(1):5-14. J Microbiol Biotechnol. 2010. PMID: 20134227 Free PMC article. Review.
Cited by
- The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota.
Leatham-Jensen MP, Frimodt-Møller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. Leatham-Jensen MP, et al. Infect Immun. 2012 May;80(5):1716-27. doi: 10.1128/IAI.06193-11. Epub 2012 Mar 5. Infect Immun. 2012. PMID: 22392928 Free PMC article. - Structure and Evolution of Glycogen Branching Enzyme N-Termini From Bacteria.
Wang L, Liu Q, Hu J, Asenso J, Wise MJ, Wu X, Ma C, Chen X, Yang J, Tang D. Wang L, et al. Front Microbiol. 2019 Jan 14;9:3354. doi: 10.3389/fmicb.2018.03354. eCollection 2018. Front Microbiol. 2019. PMID: 30692986 Free PMC article. - Genotype and phenotypes of an intestine-adapted Escherichia coli K-12 mutant selected by animal passage for superior colonization.
Fabich AJ, Leatham MP, Grissom JE, Wiley G, Lai H, Najar F, Roe BA, Cohen PS, Conway T. Fabich AJ, et al. Infect Immun. 2011 Jun;79(6):2430-9. doi: 10.1128/IAI.01199-10. Epub 2011 Mar 21. Infect Immun. 2011. PMID: 21422176 Free PMC article. - Hepatic histopathology and apoptosis in diet-induced-obese mice under Escherichia coli pneumonia.
Song H, Zuo Z, Yang Z, Gao C, Chen K, Fang J, Cui H, Ouyang P, Deng J, Geng Y, Guo H. Song H, et al. Aging (Albany NY). 2019 May 14;11(9):2836-2851. doi: 10.18632/aging.101956. Aging (Albany NY). 2019. PMID: 31085802 Free PMC article. - Genome-scale metabolic reconstruction and in silico analysis of the rice leaf blight pathogen, Xanthomonas oryzae.
Koduru L, Kim HY, Lakshmanan M, Mohanty B, Lee YQ, Lee CH, Lee DY. Koduru L, et al. Mol Plant Pathol. 2020 Apr;21(4):527-540. doi: 10.1111/mpp.12914. Epub 2020 Feb 18. Mol Plant Pathol. 2020. PMID: 32068953 Free PMC article.
References
- Alonso-Casajus, N., D. Dauvillee, A. M. Viale, F. J. Munoz, E. Baroja-Fernandez, M. T. Moran-Zorzano, G. Eydallin, S. Ball, and J. Pozueta-Romero. 2006. Glycogen phosphorylase, the product of the glgP Gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J. Bacteriol. 1885266-5272. - PMC - PubMed
- Amaretti, A., E. Tamburini, T. Bernardi, A. Pompei, S. Zanoni, G. Vaccari, D. Matteuzzi, and M. Rossi. 2006. Substrate preference of Bifidobacterium adolescentis MB 239: compared growth on single and mixed carbohydrates. Appl. Microbiol. Biotechnol. 73654-662. - PubMed
- Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. Garcia-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 19131-36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical