Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos) - PubMed (original) (raw)
Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos)
Cristina E Valdiosera et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The endangered brown bear populations (Ursus arctos) in Iberia have been suggested to be the last fragments of the brown bear population that served as recolonization stock for large parts of Europe during the Pleistocene. Conservation efforts are intense, and results are closely monitored. However, the efforts are based on the assumption that the Iberian bears are a unique unit that has evolved locally for an extended period. We have sequenced mitochondrial DNA (mtDNA) from ancient Iberian bear remains and analyzed them as a serial dataset, monitoring changes in diversity and occurrence of European haplogroups over time. Using these data, we show that the Iberian bear population has experienced a dynamic, recent evolutionary history. Not only has the population undergone mitochondrial gene flow from other European brown bears, but the effective population size also has fluctuated substantially. We conclude that the Iberian bear population has been a fluid evolutionary unit, developed by gene flow from other populations and population bottlenecks, far from being in genetic equilibrium or isolated from other brown bear populations. Thus, the current situation is highly unusual and the population may in fact be isolated for the first time in its history.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
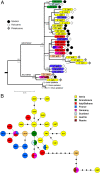
Fig. 1.
Past and current genetic diversity. (A) Bayesian phylogeny generated by using ancient and modern brown bear European sequences (long dataset) and three cave bears as outgroup. Bootstrap values are shown above the node, and posterior probabilities are shown under the nodes. The color of each square represents the geographic origin, the number in italics indicates the haplotype (see
SI Dataset 2
), and the “n” is the haplotype frequency. (B) Minimum spanning network showing the haplotype distribution of ancient and modern brown bear sequences (short dataset). Numbers inside the circles indicate the assigned haplotype from a total of 29 haplotypes obtained.
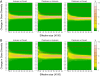
Fig. 2.
Changes in gene diversity simulated between different time periods assuming a constant population size. Mutation rate: 29.8% substitution per site per million years (A) and 10.0% substitution per site per million years (B). The colors refer to the proportion of simulations leading to the corresponding shift in gene diversity. Thirteen possible effective sizes were simulated (100, 500, 1,000, 2,000, 5,000, 10,000, 15,000, 20,000, 25,000, 30,000, 35,000, 40,000, and 45,000 individuals). Red line indicates observed changes in gene diversity (large dataset).
Similar articles
- Paths for colonization or exodus? New insights from the brown bear (Ursus arctos) population of the Cantabrian Mountains.
Gregório I, Barros T, Pando D, Morante J, Fonseca C, Ferreira E. Gregório I, et al. PLoS One. 2020 Jan 31;15(1):e0227302. doi: 10.1371/journal.pone.0227302. eCollection 2020. PLoS One. 2020. PMID: 32004321 Free PMC article. - Mitogenetic structure of brown bears (Ursus arctos L.) in northeastern Europe and a new time frame for the formation of European brown bear lineages.
Saarma U, Ho SY, Pybus OG, Kaljuste M, Tumanov IL, Kojola I, Vorobiev AA, Markov NI, Saveljev AP, Valdmann H, Lyapunova EA, Abramov AV, Männil P, Korsten M, Vulla E, Pazetnov SV, Pazetnov VS, Putchkovskiy SV, Rõkov AM. Saarma U, et al. Mol Ecol. 2007 Jan;16(2):401-13. doi: 10.1111/j.1365-294X.2006.03130.x. Mol Ecol. 2007. PMID: 17217353 - Canine adenovirus type 1 (CAdV-1) in free-ranging European brown bear (Ursus arctos arctos): A threat for Cantabrian population?
García Marín JF, Royo LJ, Oleaga A, Gayo E, Alarcia O, Pinto D, Martínez IZ, González P, Balsera R, Marcos JL, Balseiro A. García Marín JF, et al. Transbound Emerg Dis. 2018 Dec;65(6):2049-2056. doi: 10.1111/tbed.13013. Epub 2018 Sep 20. Transbound Emerg Dis. 2018. PMID: 30179311 - Impacts of Human Recreation on Brown Bears (Ursus arctos): A Review and New Management Tool.
Fortin JK, Rode KD, Hilderbrand GV, Wilder J, Farley S, Jorgensen C, Marcot BG. Fortin JK, et al. PLoS One. 2016 Jan 5;11(1):e0141983. doi: 10.1371/journal.pone.0141983. eCollection 2016. PLoS One. 2016. PMID: 26731652 Free PMC article. Review. - Sperm cryopreservation in brown bear (Ursus arctos): preliminary aspects.
Anel L, Alvarez M, Martínez-Pastor F, Gomes S, Nicolás M, Mata M, Martínez AF, Borragán S, Anel E, de Paz P. Anel L, et al. Reprod Domest Anim. 2008 Oct;43 Suppl 4:9-17. doi: 10.1111/j.1439-0531.2008.01248.x. Reprod Domest Anim. 2008. PMID: 18803752 Review.
Cited by
- Diversity lost: are all Holarctic large mammal species just relict populations?
Hofreiter M, Barnes I. Hofreiter M, et al. BMC Biol. 2010 Apr 21;8:46. doi: 10.1186/1741-7007-8-46. BMC Biol. 2010. PMID: 20409351 Free PMC article. - Paths for colonization or exodus? New insights from the brown bear (Ursus arctos) population of the Cantabrian Mountains.
Gregório I, Barros T, Pando D, Morante J, Fonseca C, Ferreira E. Gregório I, et al. PLoS One. 2020 Jan 31;15(1):e0227302. doi: 10.1371/journal.pone.0227302. eCollection 2020. PLoS One. 2020. PMID: 32004321 Free PMC article. - Genetic analyses reveal population structure and recent decline in leopards (Panthera pardus fusca) across the Indian subcontinent.
Bhatt S, Biswas S, Karanth K, Pandav B, Mondol S. Bhatt S, et al. PeerJ. 2020 Feb 4;8:e8482. doi: 10.7717/peerj.8482. eCollection 2020. PeerJ. 2020. PMID: 32117616 Free PMC article. - Loss of Mitochondrial Genetic Diversity despite Population Growth: The Legacy of Past Wolf Population Declines.
Salado I, Preick M, Lupiáñez-Corpas N, Fernández-Gil A, Vilà C, Hofreiter M, Leonard JA. Salado I, et al. Genes (Basel). 2022 Dec 26;14(1):75. doi: 10.3390/genes14010075. Genes (Basel). 2022. PMID: 36672816 Free PMC article. - Bone accumulation by leopards in the Late Pleistocene in the Moncayo massif (Zaragoza, NE Spain).
Sauqué V, Rabal-Garcés R, Sola-Almagro C, Cuenca-Bescós G. Sauqué V, et al. PLoS One. 2014 Mar 18;9(3):e92144. doi: 10.1371/journal.pone.0092144. eCollection 2014. PLoS One. 2014. PMID: 24642667 Free PMC article.
References
- Servheen C. Status and conservation of the bears of the world. Proceedings of the 8th International Conference on Bear Research and Management; British Columbia, Canada. 1990. Monograph Series No. 2.
- Mattson DJ, Merrill T. Extirpations of grizzly bears in the contiguous United States, 1850–2000. Cons Biol. 2002;16:1123–1136.
- Waits LP, Talbot SL, Ward RH, Shields GF. Phylogeography of the North American brown bear and implications for conservation. Cons Biol. 1998;12:408–417.
- Taberlet P, Bouvet J. Mitochondrial DNA polymorphism, phylogeography, and conservation genetics of the brown bear (Ursus arctos) in Europe. Proc Biol Sci. 1994;255:195–200. - PubMed
- Randi E. Conservation genetics of carnivores in Italy. Comptes Rendus Biol. 2003;326(Suppl 1):S54–S60. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources