Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool - PubMed (original) (raw)
Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool
N Spassky et al. Dev Biol. 2008.
Abstract
Cerebellar granule cell precursors (GCPs), which give rise to the most abundant neuronal type in the mammalian brain, arise from a restricted pool of primary progenitors in the rhombic lip (RL). Sonic hedgehog (Shh) secreted by developing Purkinje cells is essential for the expansion of GCPs and for cerebellar morphogenesis. Recent studies have shown that the primary cilium concentrates components of Shh signaling and that this structure is required for Shh signaling. GCPs have a primary cilium on their surface [Del Cerro, M.P., Snider, R.S. (1972). Studies on the developing cerebellum. II. The ultrastructure of the external granular layer. J Comp Neurol 144, 131-64.]. Here, we show that 1) this cilium can be conditionally ablated by crossing Kif3a(fl/-) mice with hGFAP-Cre mice, 2) removal of Kif3a from GCPs disrupts cerebellar development, and 3) these defects are due to a drastic reduction in Shh-dependent expansion of GCPs. A similar phenotype is observed when Smoothened (Smo), an essential transducer of Shh signaling, is removed from the same population of GCPs. Interestingly, Kif3a-Smo double conditional mutants show that Kif3a is epistatic to Smo. This work shows that Kif3a is essential for Shh-dependent expansion of cerebellar progenitors. Dysfunctional cilia are associated with diverse human disorders including Bardet-Biedl and Joubert syndromes. Cerebellar abnormalities observed in these patients could be explained by defects in Shh-induced GCP expansion.
Figures
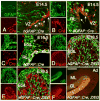
Fig. 1. The granule cell lineage is recombined by the hGFAP∷Cre transgene
(A-F) Sagittal sections of the cerebellum from a hGFAP∷Cre (A-B), hGFAP∷Cre; Z/EG (C-E) and hGFAP∷Cre; R26R (F) mice at E14.5 (A-B), E16.5 (C), E18.5 (D-E) and adult (Ad, F) stained with the antibodies shown in each panel. (A, B) Cre immunoreactivity is detected in proliferating GFAP+ cells in the ventricular zone (VZ) and the RL (arrows in A, B). (C) At E16.5, Cre+ cells are still present in the ventricular zone and in the RL but are not observed in the EGL, which already contains progeny of Cre+ cells (GFP+). (D, E) At E18.5, Cells derived from Cre+ cells (GFP+) are localized in the EGL and inner layers, and express the GCP marker Pax6 (D), but not the Purkinje cell marker CaBP (E). (F) In the adult, Bergmann glial fibers in the ML and α6+ granule cells are β-galactosidase+. Scale bars: 50 μm (A, B), 30 μm (C), 100 μm (D, E) and 300 μm (F).
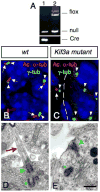
Fig.2. Lack of Kif3a inhibits ciliogenesis in the EGL
(A) PCR products obtained after amplification of genomic DNA of purified GCPs from hGFAP∷Cre; Kif3afl/- (lane 1) and Kif3afl/- (lane 2) cerebellum. Note that the LoxP band is absent in purified GCPs from hGFAP∷Cre+ mice (lane 1). (B, C) Confocal projections of double immunostaining with an anti-acetylated α-tubulin antibody (red, arrow) and an anti-γ-tubulin antibody (green, arrowhead) on sections of wild type (B) and conditional Kif3a mutant (C) at E18.5. In control animals, most GCPs extend a primary cilium (arrows) from a basal body (arrowheads). These cilia were absent in the EGL in the conditional Kif3a mutant. Basal bodies are still present in the mutant EGL (arrowhead). Note that the mutation does not affect meningeal cells since ciliary staining could still be observed in this structure (arrow in C). (D, E) Electron micrographs of GCPs from wild type and conditional Kif3a mutants at P0. (D) In wild type animals, a primary cilium (red arrow) extends from a basal body (green arrowhead) nearby a centriole (green asterisk). (E) In conditional Kif3a mutant a basal body (green arrowhead) is attached to the cell membrane nearby a centriole (green asterisk), but the primary cilium is absent. 5 μm (B-C), 0.5 μm (D-E).
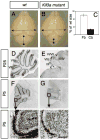
Fig. 3. Lack of Kif3a severely disrupts cerebellar development
(A-B) Gross appearance of a brain from a wild-type littermate (A) and a conditional Kif3a mutant (B) at P25. (C) The size of the forebrain (Fb) and cerebellum (Cb) from wild-type and conditional Kif3a mutant were measured as indicated by arrows in A, B. Although the forebrain from conditional Kif3a mutants is not significantly different from wild-type, the size of the cerebellum is dramatically reduced. (D-I) Cresyl violet staining of sagittal sections of cerebellum from wild type (D, F, H) and conditional Kif3a mutant (E, G, I) cerebellum at P25 (D-E) and P5 (F-I). (F-I) Lack of Kif3a limits the expansion of the EGL. (H-I) High magnifications of the squares shown in (F,G) illustrate that EGL is thinner in the mutants (red line). Scale bars: 3mm (A, B), 800 μm (D-G), 130 μm (H, I).
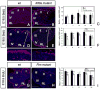
Fig. 4. Abnormal proliferation of GCPs in conditional Kif3a and Ftm mutant mice
BrdU staining of a control (A, D, G), a conditional Kif3a mutant (B, E) and a Ftm mutant (H) cerebellum at E16.5 (A-B) and E18.5 (D-E, G-H). Rostral is to the right. Squares in D, E are shown at higher magnifications below. (C, F, I) Quantification of BrdU+ cells per 1000 μm2 in regions I to IV and in the RL. Note that at E18.5, the number of BrdU+ cells is much smaller in the rostral part of the cerebellum in conditional Kif3a and Ftm mutants compared to wild type, while no significant difference was observed in caudal regions of conditional Kif3a and Ftm mutants. The BrdU+ cells were counted from at least three sections from each mouse at comparable mediolateral levels. Data from three mice per group were pooled for statistical analysis with Student’s _t_-test. **: p<0.01. Scale bars: 100 μm (A, B), 200 μm (D-E, G-H).
Fig. 5. Abnormal expression of Shh and Shh target genes, Gli1 and cyclin D1, in conditional Kif3a mutants
In situ hybridization on sagittal sections of wild type (A, D) and mutant (B, E) cerebellum at P2 with antisense riboprobes specific for Shh (A-B) and Gli1 (D-E). The location of the EGL and PCL are indicated on each panel. Note that whereas Shh expression is increased in the PCL of mutant compared to wild-type (A-C), Gli1 was no longer detected neither in the EGL nor the PCL in the mutant (E). (C, F-G) Levels of Shh mRNAs in the large cell fraction enriched in Purkinje cells (C), and levels of Gli1 (F) and cyclin D1 mRNAs in GCPs (G) at P2-4 were evaluated by semi-quantitative real-time RT-PCR and are shown as the relative quantities normalized to the level of hprt mRNA expression. The level of Shh mRNA in conditional Kif3a mutant is significantly higher than that in the wild-type, whereas the levels of Gli1 and cyclin D1 mRNAs in the mutant are significantly lower than those in the wild-type. Each column and the vertical line represent the mean± SEM. Data from three mice per group were pooled for statistical analysis with Student’s _t_-test. *: p<0.05; **: p<0.01. Scale bar: 80 μm.
Fig. 6. Abnormal Shh-induced proliferation of GCPs in cerebellar cultures from conditional Kif3a mutants
BrdU staining on cerebella sagittal slices from P3 wild type (A, C) and mutant mice (B, D) cultured with no stimulus (A-B) or 3 μg/ml Shh-N (C-D). BrdU staining (green) was counterstained with DAPI (blue) and visualized using confocal microscopy. (E) Quantification of the percentage of BrdU+ and DAPI+ nuclei in wild type and conditional Kif3a mutant slices with and without Shh-N. (F-G) Proliferation of isolated GCPs in aggregates obtained from P1 Kif3a mutants (G) compared to controls (F) with Shh-N for 48h. (H) Quantification of aggregate radius in wild type and conditional Kif3a mutants after 48h. Data from three mice per group were pooled for statistical analysis with Student’s _t_-test. ***: p<0.001. Scale bar: 100 μm.
Fig.7. Abnormal proliferation of GCPs in conditional Smo mutant and conditional Kif3a and Smo double mutant mice
(A-C) Cresyl violet staining of sagittal sections of cerebellum from conditional Kif3a mutant (A), conditional Smo mutant (B), and conditional Kif3a and Smo double mutant cerebellum at P25. Rostral is to the right. (D) Double immunostaining with an anti-acetylated α-tubulin antibody (red) and an anti-γ-tubulin antibody (green) on sections of conditional Smo mutant at E18.5. In conditional Smo mutants, most GCPs extend a primary cilium (arrow) from a basal body. Each nucleus associated with a primary cilium is indicated by an asterisk. (E-F) BrdU staining of a conditional Smo mutant (E) and a conditional Kif3a and Smo double mutant (F) cerebellum at E18.5. Rostral is to the right. (G) Quantification of BrdU+ cells per 1000 μm2 in regions I to IV and in the RL. Note that the number of BrdU+ cells is much smaller in conditional Smo mutants than in conditional Kif3a and Smo double mutants, while no significant difference was observed between conditional Kif3a mutants and conditional Kif3a and Smo double mutants (compare with Fig.4F). The BrdU+ cells were counted from at least three sections from each mouse at comparable mediolateral levels. Data from three mice per group were pooled for statistical analysis with Student’s _t_-test. **: p<0.01. Scale bar: 230 μm (A-C), 2 μm (D) and 100 μm (E-F).
Similar articles
- Multifaceted Functions of Rab23 on Primary Cilium-Mediated and Hedgehog Signaling-Mediated Cerebellar Granule Cell Proliferation.
Hor CHH, Lo JCW, Cham ALS, Leong WY, Goh ELK. Hor CHH, et al. J Neurosci. 2021 Aug 11;41(32):6850-6863. doi: 10.1523/JNEUROSCI.3005-20.2021. Epub 2021 Jul 1. J Neurosci. 2021. PMID: 34210780 Free PMC article. - Genetic deletion of genes in the cerebellar rhombic lip lineage can stimulate compensation through adaptive reprogramming of ventricular zone-derived progenitors.
Wojcinski A, Morabito M, Lawton AK, Stephen DN, Joyner AL. Wojcinski A, et al. Neural Dev. 2019 Feb 14;14(1):4. doi: 10.1186/s13064-019-0128-y. Neural Dev. 2019. PMID: 30764875 Free PMC article. - Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool.
Chizhikov VV, Davenport J, Zhang Q, Shih EK, Cabello OA, Fuchs JL, Yoder BK, Millen KJ. Chizhikov VV, et al. J Neurosci. 2007 Sep 5;27(36):9780-9. doi: 10.1523/JNEUROSCI.5586-06.2007. J Neurosci. 2007. PMID: 17804638 Free PMC article. - Sonic hedgehog patterning during cerebellar development.
De Luca A, Cerrato V, Fucà E, Parmigiani E, Buffo A, Leto K. De Luca A, et al. Cell Mol Life Sci. 2016 Jan;73(2):291-303. doi: 10.1007/s00018-015-2065-1. Epub 2015 Oct 24. Cell Mol Life Sci. 2016. PMID: 26499980 Free PMC article. Review. - Endocytic Control of Cellular Signaling at the Primary Cilium.
Pedersen LB, Mogensen JB, Christensen ST. Pedersen LB, et al. Trends Biochem Sci. 2016 Sep;41(9):784-797. doi: 10.1016/j.tibs.2016.06.002. Epub 2016 Jun 27. Trends Biochem Sci. 2016. PMID: 27364476 Review.
Cited by
- Developmental and regenerative paradigms of cilia regulated hedgehog signaling.
Kopinke D, Norris AM, Mukhopadhyay S. Kopinke D, et al. Semin Cell Dev Biol. 2021 Feb;110:89-103. doi: 10.1016/j.semcdb.2020.05.029. Epub 2020 Jun 12. Semin Cell Dev Biol. 2021. PMID: 32540122 Free PMC article. Review. - Rac-deficient cerebellar granule neurons die before they migrate to the internal granule layer.
Katayama KI, Zheng Y, Inoue N. Katayama KI, et al. Sci Rep. 2022 Sep 1;12(1):14848. doi: 10.1038/s41598-022-19252-y. Sci Rep. 2022. PMID: 36050459 Free PMC article. - Analysis of human samples reveals impaired SHH-dependent cerebellar development in Joubert syndrome/Meckel syndrome.
Aguilar A, Meunier A, Strehl L, Martinovic J, Bonniere M, Attie-Bitach T, Encha-Razavi F, Spassky N. Aguilar A, et al. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16951-6. doi: 10.1073/pnas.1201408109. Epub 2012 Oct 1. Proc Natl Acad Sci U S A. 2012. PMID: 23027964 Free PMC article. - Arborization of dendrites by developing neocortical neurons is dependent on primary cilia and type 3 adenylyl cyclase.
Guadiana SM, Semple-Rowland S, Daroszewski D, Madorsky I, Breunig JJ, Mykytyn K, Sarkisian MR. Guadiana SM, et al. J Neurosci. 2013 Feb 6;33(6):2626-38. doi: 10.1523/JNEUROSCI.2906-12.2013. J Neurosci. 2013. PMID: 23392690 Free PMC article. - Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex.
Wang L, Hou S, Han YG. Wang L, et al. Nat Neurosci. 2016 Jul;19(7):888-96. doi: 10.1038/nn.4307. Epub 2016 May 23. Nat Neurosci. 2016. PMID: 27214567 Free PMC article.
References
- Altman J, Bayer SA. Development of the Cerebellar system: In relation to its evolution, Structure, and Functions. New York, NY: CRC Press; 1997.
- Arts HH, Doherty D, van Beersum SE, Parisi MA, Letteboer SJ, Gorden NT, Peters TA, Marker T, Voesenek K, Kartono A, Ozyurek H, Farin FM, Kroes HY, Wolfrum U, Brunner HG, Cremers FP, Glass IA, Knoers NV, Roepman R. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39:882–8. - PubMed
- Baptista CA, Hatten ME, Blazeski R, Mason CA. Cell-cell interactions influence survival and differentiation of purified Purkinje cells in vitro. Neuron. 1994;12(2):243–60. - PubMed
- Bignami A, Eng LF, Dahl D, Uyeda CT. Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Res. 1972;43:429–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK063720/DK/NIDDK NIH HHS/United States
- R01 NS028478/NS/NINDS NIH HHS/United States
- R37 NS028478/NS/NINDS NIH HHS/United States
- NS 028478/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous