Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition - PubMed (original) (raw)
Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition
Sviatoslav N Bagriantsev et al. Mol Biol Cell. 2008 Jun.
Abstract
The [PSI(+)] prion is the aggregated self-propagating form of the Sup35 protein from the yeast Saccharomyces cerevisiae. Aggregates of Sup35 in [PSI(+)] cells exist in different heritable conformations, called "variants," and they are composed of detergent-resistant Sup35 polymers, which may be closely associated with themselves, other proteins, or both. Here, we report that disassembly of the aggregates into individual Sup35 polymers and non-Sup35 components increases their infectivity while retaining their variant specificity, showing that variant-specific [PSI(+)] infection can be transmitted by Sup35 polymers alone. Morphological analysis revealed that Sup35 isolated from [PSI(+)] yeast has the appearance of short barrels, and bundles, which seem to be composed of barrels. We show that the major components of two different variants of [PSI(+)] are interacting infectious Sup35 polymers and Ssa1/2. Using a candidate approach, we detected Hsp104, Ssb1/2, Sis1, Sse1, Ydj1, and Sla2 among minor components of the aggregates. We demonstrate that Ssa1/2 efficiently binds to the prion domain of Sup35 in [PSI(+)] cells, but that it interacts poorly with the nonaggregated Sup35 found in [psi(-)] cells. Hsp104, Sis1, and Sse1 interact preferentially with the prion versus nonprion form of Sup35, whereas Sla2 and Ssb1/2 interact with both forms of Sup35 with similar efficiency.
Figures
Figure 1.
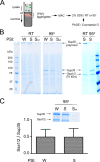
Sup35 and Ssa1/2 are the major components of weak and strong [PSI+] aggregates. (A) Experimental scheme. Isolation of Sup35 (expressed from a centromeric vector under the SUP35 promoter) and associated proteins from [_pin_−] _sup35_Δ yeast was facilitated by a his6 tag on Sup35 (see Materials and Methods). Yeast lysates were fractionated by centrifugation (c/f) on a sucrose cushion, and the fraction containing heavy protein aggregates was collected and subjected to metal affinity chromatography (MAC). Proteins were eluted from the MAC column, treated with SDS at room temperature (RT) or at 95°C, and then analyzed by SDS-PAGE and Coomassie G250. (B) Ssa1/2 coisolated with his6-tagged Sup35 from W and S [PSI+] lysates. The levels of proteins that were not specifically associated with Sup35, i.e., that also occurred in the no-his6 tag [PSI+] control (Swt), varied between isolations and sometimes were minimal (right panel). Sup35 polymers were resistant to SDS treatment at RT (seen in the bottom of the loading wells), but they disassembled into monomers at 95°C. Ssa1/2 was monomeric at both temperatures. (C) Ssa1/2 was on average two times less abundant than Sup35 in W and S [PSI+] aggregates. The relative abundance of the Coomassie G-stained Sup35 and Ssa1/2 was measured by an in-gel densitometry. The assumption was made that the dye binds with equal efficacy to both proteins. The intensities of the corresponding areas in the control Swt [PSI+] eluate were subtracted from the initial values for Ssa and Sup35 in W and S [PSI+] eluates before calculation. Data are presented as mean ± SD, n = 3. The strains used are [_pin_−][PSI+]: L2988 (W), L2982 (S), L2979 (Swt).
Figure 2.
Immunoblot analysis of major and minor protein components of weak and strong [PSI+] aggregates. [PSI+] aggregates were obtained as described in Figure 1 (eluates), and then they were analyzed by immunoblotting with the indicated antibodies along with initial lysates after treatment with SDS at RT or at 95°C. (A) SDS treatment of W [PSI+] lysates and eluates at room temperature revealed some Sup35 monomer, which was almost undetectable in S [PSI+] samples. (B) In addition to Sup35 and Ssa1/2 (major components shown in A), [PSI+] aggregates (eluates) contained Sla2, Hsp104, Sse1, Ssb1/2, Ydj1, and Sis1 (minor components). Rnq1 did not coisolate with Sup35.
Figure 3.
Ssa1/2 efficiently interacts with the prion but not the nonprion form of Sup35N(1-137). Interaction between Sup35 and Ssa1/2 was assayed in [PSI+] (weak or strong variant) or [_psi_−] strains by immunocapturing (IC) Sup35 or Ssa1/2 from whole unfractionated lysates (equalized by total protein) with α-his6 or α-Ssa1/2 antibody, respectively, immobilized on protein G-coupled magnetic beads (see Materials and Methods). After incubation, the beads were washed and the specifically bound proteins were eluted at 95°C in an SDS-containing buffer and analyzed by immunoblotting (IB) with indicated antibodies. Lysates of the same strains incubated along with the experimental samples without antibodies (lysates before IC) showed no degradation of Sup35 or Ssa1/2. The following strains were used: _sup35_Δ expressing Sup35-his6 (A) and SUP35 yeast without plasmid (B). (C) _sup35_Δ2-252 yeast expressing the 137-amino acid-long N-terminal fragment of Sup35 (Sup35N1-137).
Figure 4.
Some components of [PSI+] aggregates interact poorly with Sup35 from [_psi_−]. Interaction between Sup35 and minor components of [PSI+] aggregates was assayed in S [PSI+] or [_psi_−] _sup35_Δ yeast expressing Sup35-HA, by immunocapture (IC) and imunoblotting (IB) as described in Figure 3. Sla2 and Ssb1/2 interacted with Sup35 in [PSI+] and [_psi_−] with similar efficiency. Hsp104, Sse1, Ssa1/2, and Sis1, preferentially interacted with Sup35 from [PSI+]. Ydj1 association with Sup35 was not detected by immunocapture, although association of Ydj1 with [PSI+] aggregates was shown by another method (see Figure 2B).
Figure 5.
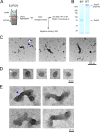
Morphological analysis of Sup35 isolated from [PSI+] yeast. (A) Experimental scheme. Purification of Sup35 polymers was performed from an S [PSI+] strain as in Figure 1, but with more stringent washing conditions (see Materials and Methods). (B) SDS-PAGE/Coomassie G250 analysis of Sup35 polymers isolated from S [PSI+] yeast in A and treated with SDS at RT or at 95°C. (C–E) Electron micrographs of Sup35 polymers negatively stained with uranyl acetate (C). The sample contained numerous barrel-shaped structures (white arrowhead, D), which appeared to frequently form bundles (blue arrowhead, E). These structures were never detected in control isolations using [_psi_−] yeast expressing Sup35-his6 or S [PSI+] yeast expressing an untagged Sup35.
Figure 6.
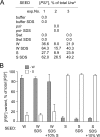
SDS-treated [PSI+] aggregates induce [PSI+] in a variant-specific manner. A [_psi_−] ade1-14 ura3-14 yeast strain was transformed with a URA3 plasmid mixed with [PSI+] aggregate preparations (SEED) obtained from the indicated strains as described in Figure 1 and treated or not with 1% SDS at room temperature. The presence of [PSI+] was tested among Ura+ transformants by using the ade1-14 nonsense suppression assay (see Materials and Methods and Supplemental Figure S1A). At least 112 Ura+ transformants were analyzed for each sample in each experiment. (A) SDS treatment increased the infectivity of [PSI+] aggregates, as judged by the increase in the percentage of [PSI+] colonies among Ura+ transformants. Data from three independent experiments are shown. (B) [PSI+] aggregates induced the same variant of [PSI+], regardless of SDS treatment. Where indicated, 10% (vol/vol) of untreated aggregates from the opposite [PSI+] variant was added to the transformation mixture. Data are presented as mean ± SD, n = 3.
Figure 7.
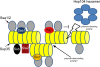
Model of the intracellular organization of [PSI+] aggregates. [PSI+] aggregates are proposed to be mainly composed of interacting infectious Sup35 polymers of various sizes, heavily decorated with Ssa1/2. The binding of Ssa1/2 to the polymers may be regulated via its cochaperones Sis1, Sse1, and Ydj1, which bind to Sup35 polymers independently or with Ssa1/2. Sup35 polymers also interact with Ssb1/2 and Sla2. Ssa1/2, which we find in an ∼1:2 ratio with Sup35, is shown shielding Sup35 from Hsp104's shearing activity to explain the previous finding that excess Ssa1/2 in [PSI+] causes an increase in Sup35 polymer size (Allen et al., 2005). The hypothetical polymer-dissociating activity, which separates individual polymers from each other, is indicated.
Similar articles
- Prion and nonprion amyloids: a comparison inspired by the yeast Sup35 protein.
Kushnirov VV, Vishnevskaya AB, Alexandrov IM, Ter-Avanesyan MD. Kushnirov VV, et al. Prion. 2007 Jul-Sep;1(3):179-84. doi: 10.4161/pri.1.3.4840. Epub 2007 Jul 6. Prion. 2007. PMID: 19164899 Free PMC article. Review. - Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104.
Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Kryndushkin DS, et al. J Biol Chem. 2003 Dec 5;278(49):49636-43. doi: 10.1074/jbc.M307996200. Epub 2003 Sep 24. J Biol Chem. 2003. PMID: 14507919 - Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions.
Shorter J, Lindquist S. Shorter J, et al. EMBO J. 2008 Oct 22;27(20):2712-24. doi: 10.1038/emboj.2008.194. Epub 2008 Oct 2. EMBO J. 2008. PMID: 18833196 Free PMC article. - Effect of charged residues in the N-domain of Sup35 protein on prion [PSI+] stability and propagation.
Bondarev SA, Shchepachev VV, Kajava AV, Zhouravleva GA. Bondarev SA, et al. J Biol Chem. 2013 Oct 4;288(40):28503-13. doi: 10.1074/jbc.M113.471805. Epub 2013 Aug 21. J Biol Chem. 2013. PMID: 23965990 Free PMC article. - The life of [PSI].
Cox B, Tuite M. Cox B, et al. Curr Genet. 2018 Feb;64(1):1-8. doi: 10.1007/s00294-017-0714-7. Epub 2017 Jun 26. Curr Genet. 2018. PMID: 28653109 Free PMC article. Review.
Cited by
- Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates.
Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Malinovska L, et al. Mol Biol Cell. 2012 Aug;23(16):3041-56. doi: 10.1091/mbc.E12-03-0194. Epub 2012 Jun 20. Mol Biol Cell. 2012. PMID: 22718905 Free PMC article. - Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system.
Nakagawa Y, Shen HC, Komi Y, Sugiyama S, Kurinomaru T, Tomabechi Y, Krayukhina E, Okamoto K, Yokoyama T, Shirouzu M, Uchiyama S, Inaba M, Niwa T, Sako Y, Taguchi H, Tanaka M. Nakagawa Y, et al. Nat Chem Biol. 2022 Mar;18(3):321-331. doi: 10.1038/s41589-021-00951-y. Epub 2022 Feb 17. Nat Chem Biol. 2022. PMID: 35177839 - Beyond Amyloid Fibers: Accumulation, Biological Relevance, and Regulation of Higher-Order Prion Architectures.
Naeimi WR, Serio TR. Naeimi WR, et al. Viruses. 2022 Jul 27;14(8):1635. doi: 10.3390/v14081635. Viruses. 2022. PMID: 35893700 Free PMC article. Review. - Variant-specific prion interactions: Complicating factors.
Sharma J, Liebman SW. Sharma J, et al. Cell Logist. 2013 Jan 1;3(1):e25698. doi: 10.4161/cl.25698. Epub 2013 Sep 12. Cell Logist. 2013. PMID: 24475372 Free PMC article. - Extracellular Vesicles and the Propagation of Yeast Prions.
Kabani M. Kabani M. Curr Top Microbiol Immunol. 2021;432:57-66. doi: 10.1007/978-3-030-83391-6_6. Curr Top Microbiol Immunol. 2021. PMID: 34972878
References
- Allen K. D., Chernova T. A., Tennant E. P., Wilkinson K. D., Chernoff Y. O. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 2007;282:3004–3013. - PubMed
- Allen K. D., Wegrzyn R. D., Chernova T. A., Muller S., Newnam G. P., Winslett P. A., Wittich K. B., Wilkinson K. D., Chernoff Y. O. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. - PMC - PubMed
- Bagriantsev S., Liebman S. W. Specificity of prion assembly in vivo: [PSI+] and [PIN+] form separate structures in yeast. J. Biol. Chem. 2004;279:51042–51048. - PubMed
- Bagriantsev S. N., Kushnirov V. V., Liebman S. W. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-56350/GM/NIGMS NIH HHS/United States
- MH-073156/MH/NIMH NIH HHS/United States
- R01 GM056350/GM/NIGMS NIH HHS/United States
- R01 MH073156-02/MH/NIMH NIH HHS/United States
- R01 MH073156/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases