Chromatin dynamics during epigenetic reprogramming in the mouse germ line - PubMed (original) (raw)
. 2008 Apr 17;452(7189):877-81.
doi: 10.1038/nature06714. Epub 2008 Mar 19.
Affiliations
- PMID: 18354397
- PMCID: PMC3847605
- DOI: 10.1038/nature06714
Chromatin dynamics during epigenetic reprogramming in the mouse germ line
Petra Hajkova et al. Nature. 2008.
Abstract
A unique feature of the germ cell lineage is the generation of totipotency. A critical event in this context is DNA demethylation and the erasure of parental imprints in mouse primordial germ cells (PGCs) on embryonic day 11.5 (E11.5) after they enter into the developing gonads. Little is yet known about the mechanism involved, except that it is apparently an active process. We have examined the associated changes in the chromatin to gain further insights into this reprogramming event. Here we show that the chromatin changes occur in two steps. The first changes in nascent PGCs at E8.5 establish a distinctive chromatin signature that is reminiscent of pluripotency. Next, when PGCs are residing in the gonads, major changes occur in nuclear architecture accompanied by an extensive erasure of several histone modifications and exchange of histone variants. Furthermore, the histone chaperones HIRA and NAP-1 (NAP111), which are implicated in histone exchange, accumulate in PGC nuclei undergoing reprogramming. We therefore suggest that the mechanism of histone replacement is critical for these chromatin rearrangements to occur. The marked chromatin changes are intimately linked with genome-wide DNA demethylation. On the basis of the timing of the observed events, we propose that if DNA demethylation entails a DNA repair-based mechanism, the evident histone replacement would represent a repair-induced response event rather than being a prerequisite.
Figures
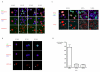
Figure 1. Following the entry of PGCs into the gonads, germ cell chromatin undergoes rapid conformational changes.
A) Dissapearance of linker histone H1 in PGCs at E11.5. PGC7/Stella (in red) was used as germ cell specific marker. PGCs are depicted by arrows. B) Increase in the nuclear size and disappearance of chromocenters in PGCs at E11.5 as observed by DAPI staining of cell suspension from genital ridges. PGCs are depicted by arrows. The lower panel shows identification of PGCs by staining with germ cell specific marker (SSEA1 or Oct4). Scale bars: 10μm. C) Measurement of the nuclear size on the cryosections from E10.5-E13.5 genital ridges shows transient change in the size of germ cell nuclei. The increase is highly statistically significant using t-test. The measured PGC nuclei were identified on the cryosections using Oct4 stainings (data not shown).
Figure 2. Chromatin changes observed in PGCs at E11.5.
A) Presence of H3K27me3 in PGCs. At E10.5 nuclei of PGCs contain high levels of H3K27me3 as part of their germline chromatin signature (for details see text). This chromatin mark is lost in PGCs at E11.5 and re-gained between E12.5 and E13.5. Notably the loss of H3K27me3 occurs despite the continuous presence of Ezh2 responsible for this modification in PGCs (data not shown). B) Loss of H3K9me3 in PGCs at E11.5. Stainings were performed on single cell suspensions from genital ridges between E10.5-E13.5. The shown epigenetic changes are detectable in 70-90% of PGCs depending on the developmental stage of the embryo. Germ cells were stained in green using the germ cell specific markers SSEA1 and Oct4, respectively, and are depicted by arrows. Scale bar: 10μm. C) The immunofluorescence stainings of cryosections of embryonic gonads (as described in Methods section). Oct4 positive germ cells (depicted by arrows) are stained in green. Note high levels of H2A.Z in early postmigratory PGCs at E10.5 and the disappearance of the H2A.Z staining in later developmental stages (E11.5 and E12.5), while the signal in surrounding somatic cells remains relatively constant. Scale bar: 10μm. D) Quantification of the H2A.Z signal in germ cells. In order to normalize between different samples, the intensity of H2A.Z staining was calculated as a mean pixel intensity in every PGC divided by the mean pixel intensity in surrounding somatic cells. The Y axis values thus represent the intensity of H2A.Z staining in PGCs in comparison with surrounding somatic cells.
Figure 3. Separation and analysis of PGCs undergoing distinct phases of the reprogramming process.
A) Transient appearance of two populations of GFP expressing PGCs at E11.5. The X and Y axis represent the FSC (forward scatter) and GFP intensity values, respectively. The lower panels show the profiles of the GFP (germ cell specific) signal. Note the appearance of two distinct peaks at E11.5. B) Separation and analysis of chromatin configuration of the two populations of PGC, denoted as A and B, at E11.5. The population B is more advanced and shows loss of histone H1 and H2A.Z. Note the cytoplasmic localisation of CAF-1 in these populations and relocalisation of NAP1 from the cytoplasm to the nucleus in population B (for details see the text). Cells shown here are representative of the populations. Between 50 and 100 PGCs were examined for each population. Scale bar: 10μm. C) Analysis of DNA methylation of peg3 and lit1 (kcnqtot1) DMRs in dinstict populations of E11.5 PGCs using bisulphite genomic sequencing. Each line represents an independent clone. Open and closed circles represent non-methylated and methylated CpGs, respectively.
Figure 4. Possible connections between chromatin changes and DNA demethylation process.
Changes in chromatin structure may be seen as prerequisite for the DNA demethylation process (accessibility model.) Alternatively, DNA demethylation in PGCs can occur via DNA repair pathway as described in plants , , . The resulting DNA damage and repair could potentially induce chromatin remodelling, which would occur as a consequence of DNA demethylation (for details see the text).
Similar articles
- Characterization of the Epigenetic Changes During Human Gonadal Primordial Germ Cells Reprogramming.
Eguizabal C, Herrera L, De Oñate L, Montserrat N, Hajkova P, Izpisua Belmonte JC. Eguizabal C, et al. Stem Cells. 2016 Sep;34(9):2418-28. doi: 10.1002/stem.2422. Epub 2016 Jun 30. Stem Cells. 2016. PMID: 27300161 Free PMC article. - Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway.
Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Hajkova P, et al. Science. 2010 Jul 2;329(5987):78-82. doi: 10.1126/science.1187945. Science. 2010. PMID: 20595612 Free PMC article. - Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice.
Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Seki Y, et al. Development. 2007 Jul;134(14):2627-38. doi: 10.1242/dev.005611. Epub 2007 Jun 13. Development. 2007. PMID: 17567665 - A current view of the epigenome in mouse primordial germ cells.
Matsui Y, Mochizuki K. Matsui Y, et al. Mol Reprod Dev. 2014 Feb;81(2):160-70. doi: 10.1002/mrd.22214. Epub 2013 Aug 7. Mol Reprod Dev. 2014. PMID: 23868517 Review. - Epigenetic profiles in primordial germ cells: global modulation and fine tuning of the epigenome for acquisition of totipotency.
Mochizuki K, Matsui Y. Mochizuki K, et al. Dev Growth Differ. 2010 Aug;52(6):517-25. doi: 10.1111/j.1440-169X.2010.01190.x. Dev Growth Differ. 2010. PMID: 20646024 Review.
Cited by
- The reciprocal relationship between primordial germ cells and pluripotent stem cells.
Pirouz M, Klimke A, Kessel M. Pirouz M, et al. J Mol Med (Berl). 2012 Jul;90(7):753-61. doi: 10.1007/s00109-012-0912-1. Epub 2012 May 15. J Mol Med (Berl). 2012. PMID: 22584374 Review. - Large, male germ cell-specific hypomethylated DNA domains with unique genomic and epigenomic features on the mouse X chromosome.
Ikeda R, Shiura H, Numata K, Sugimoto M, Kondo M, Mise N, Suzuki M, Greally JM, Abe K. Ikeda R, et al. DNA Res. 2013 Dec;20(6):549-65. doi: 10.1093/dnares/dst030. Epub 2013 Jul 15. DNA Res. 2013. PMID: 23861320 Free PMC article. - Regenerative Medicine Approaches in Bioengineering Female Reproductive Tissues.
Sittadjody S, Criswell T, Jackson JD, Atala A, Yoo JJ. Sittadjody S, et al. Reprod Sci. 2021 Jun;28(6):1573-1595. doi: 10.1007/s43032-021-00548-9. Epub 2021 Apr 20. Reprod Sci. 2021. PMID: 33877644 Review. - Epigenetics and developmental programming of adult onset diseases.
O'Sullivan L, Combes AN, Moritz KM. O'Sullivan L, et al. Pediatr Nephrol. 2012 Dec;27(12):2175-82. doi: 10.1007/s00467-012-2108-x. Pediatr Nephrol. 2012. PMID: 22302599 Review. - Specialty Grand Challenge-Assisted Reproduction.
Lunenfeld E. Lunenfeld E. Front Reprod Health. 2021 Jul 26;3:551499. doi: 10.3389/frph.2021.551499. eCollection 2021. Front Reprod Health. 2021. PMID: 36304062 Free PMC article. No abstract available.
References
- Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. - PubMed
- Lee J, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–17. - PubMed
- Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–13. - PubMed
- Ancelin K, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials