An extended transcriptional network for pluripotency of embryonic stem cells - PubMed (original) (raw)
An extended transcriptional network for pluripotency of embryonic stem cells
Jonghwan Kim et al. Cell. 2008.
Erratum in
- Cell. 2008 Jun 27;133(7):1290
Abstract
Much attention has focused on a small set of transcription factors that maintain human or mouse embryonic stem (ES) cells in a pluripotent state. To gain a more complete understanding of the regulatory network that maintains this state, we identified target promoters of nine transcription factors, including somatic cell reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) and others (Nanog, Dax1, Rex1, Zpf281, and Nac1), on a global scale in mouse ES cells. We found that target genes fall into two classes: promoters bound by few factors tend to be inactive or repressed, whereas promoters bound by more than four factors are largely active in the pluripotent state and become repressed upon differentiation. Furthermore, we propose a transcriptional hierarchy for reprogramming factors and broadly distinguish targets of c-Myc versus other factors. Our data provide a resource for exploration of the complex network maintaining pluripotency.
Figures
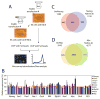
Figure 1. Strategy of in vivo biotinylation-mediated chromatin immunoprecipitation and microarray (bioChIP-chip)
(A) Schematic representation of biotin-mediated chromatin immunoprecipitation. The gray bar represents BirA target sequence (MSGLNDIFEAQKIEWHEGAPSSR). (B) Expression analysis of nine genes using cell lines expressing biotin-tagged proteins. Biotin-tagged cell lines are indicated on the horizontal axis and transcript levels are presented as color bars. (C)–(D) Overlap of target promoters between bioChIP-chip and conventional ChIP-chip experiments for Nanog (C) and Myc (D). Predicted overlap might be underestimated due to a fixed statistical threshold (see also Figure 2, and Figure S3).
Figure 2. Chromosomal view of Nanog occupancy detected by bioChIP-chip and conventional ChIP-chip
(A) Comparison of Nanog binding patterns using multiple cell lines is displayed using Affymetrix Integrated Genome Browser. In addition to bioNanog ChIP (top), antibody ChIP-chip data from control cell lines (J1 ES and BirA expressing cells) and cells expressing ectopic biotin-tagged protein (bioDax1 and bioOct4 cells) are tested. Non-specific biotinylation by BirA enzyme was also tested (bottom). Yellow box indicates the chromosomal loci harboring Gbx1. (B) Representative view of Nanog occupancy to its target Gbx1 upstream promoter.
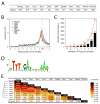
Figure 3. Summary of nine transcription factor occupancy and histone modification status
(A) Number of target promoters bound by each factor or associated with H3K4 or H3K27 trimethylation. (B) Relative position of chromosomal target loci of each factor to the TSS. (C) Number of common targets of multiple factors. Y-axis represents the number of target promoters occupied by transcription factor(s). Red dots represent the accumulated number of target promoters. (D) Predicted consensus binding motif of multiple factor target loci using MEME. (E) Correlation between each factor targets and hierarchical cluster of nine factors based on their target similarity.
Figure 4. H3K4me3 and H3K27me3 status and factor occupancy of the promoters
(A) A supervised cluster image showing 6632 target promoters occupied by different factor combinations (see Experimental Procedure). Corresponding H3K4me3 (red) and H3K27me3 (blue) histone marks (presence: 1; absence: 0) as well as gene expression profiles (log2) upon J1 ES cell differentiation (0–18h: red, 4–14d: blue, see Experimental Procedure) are shown as moving window averaged lines (bin size 50 and step size 1). Bar ‘a’ represents the promoters occupied by multiple factors including at least Nanog, Sox2, Dax1, Nac1 and Oct4 (left panel) and corresponding gene expression changes upon differentiation (middle panel) as well as their histone marks (right panel). Bars ‘b’ represent the clusters of promoters occupied by a single factor Nanog, Dax1, Klf4, and Zfp281 respectively (see also Figure 5E and 5H). Green lines (bars c) represent Myc target promoters with corresponding gene expression profiles and histone mark status. (B) H3K4me3 (red line) and H3K27me3 (blue line) status for Myc target promoters. (C) Expression profiles of Myc target genes at different time points upon differentiation (0–18h: red, 4–14d: blue). Total 6632 target genes of any of 9 factors are shown, and moving window average (bin size 50 and step size 1) was applied (B–C). (D) Factor target promoters are both H3K4me3 and H3K27me3 rich over all promoters. ‘7TFs’ represent the targets of any of 7 factors (Nanog, Sox2, Dax1, Nac1, Oct4, Klf4, Zfp281), and ‘All’ represents all promoters. Asterisk indicates hypergeometiric probability < 0.0001. (E) Histone marks on the target promoters of each factor. Asterisk indicates p-value < 0.0001.
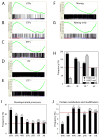
Figure 5. Target gene expression and transcription factor occupancy on their promoters
(A)–(E) GSEA analyses showing the relationship between target gene expression and factor occupancy. Target promoters were classified based on the number of co-occupying factors and corresponding gene expression upon differentiation was tested. Common targets of 6 factors (A) are enriched in active genes in ES cells, whereas single factor only targets are more repressed (D). ‘1TF*’ represents a subset of ‘1TF’ which includes promoters solely occupied by either Nanog, Dax1, Klf4 or Zfp281 as described in Figure 4A, bars b (E). (F)–(G) Nanog targets are both active and repressed in ES cells (F), however targets only occupied by Nanog are repressed (G). (H) Common target promoters of 6 factors (Nanog, Sox2, Dax1, Nac1, Oct4 and Klf4) are enriched for H3K4me3 marks and reduced for H3K27me3 marks. Promoters occupied by only one factor show an increase in H3K27me3 marks (1TF and 1TF*). Double asterisk indicates p-value < 0.0001, and single asterisk indicates p-value = 0.006. (I)–(J) Genes of multiple factor targets (at least 4TFs) are enriched in developmental processes.
Figure 6. Expanded transcriptional regulatory network and regulatory circuit within 4 somatic cell reprogramming factors and Nanog
(A) Transcriptional regulatory circuit within nine factors. Five factors (Nanog, Oct4, Sox2, Dax1, and Klf4) show auto-regulatory mechanism. (B) Expanded transcriptional regulatory network showing target hubs of multiple factors within the previously identified protein interaction network in ES cells. Yellow circles represent nine factors we examined. The size of each circle reflects the degree of factor co-occupancy. Arrowhead indicates the direction of transcriptional regulation (A–B). Sox2, Klf4 and Myc were not in the original protein interaction network (Wang et al., 2006). (C). Transcriptional regulatory circuit within 4 somatic cell reprogramming factors and Nanog
Similar articles
- Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells.
Kidder BL, Yang J, Palmer S. Kidder BL, et al. PLoS One. 2008;3(12):e3932. doi: 10.1371/journal.pone.0003932. Epub 2008 Dec 11. PLoS One. 2008. PMID: 19079543 Free PMC article. - Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells.
Yang J, Chai L, Fowles TC, Alipio Z, Xu D, Fink LM, Ward DC, Ma Y. Yang J, et al. Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19756-61. doi: 10.1073/pnas.0809321105. Epub 2008 Dec 5. Proc Natl Acad Sci U S A. 2008. PMID: 19060217 Free PMC article. - Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells.
Liu X, Huang J, Chen T, Wang Y, Xin S, Li J, Pei G, Kang J. Liu X, et al. Cell Res. 2008 Dec;18(12):1177-89. doi: 10.1038/cr.2008.309. Cell Res. 2008. PMID: 19030024 - The transcriptional network controlling pluripotency in ES cells.
Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN. Orkin SH, et al. Cold Spring Harb Symp Quant Biol. 2008;73:195-202. doi: 10.1101/sqb.2008.72.001. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19478325 Review. - Directing reprogramming to pluripotency by transcription factors.
Adachi K, Schöler HR. Adachi K, et al. Curr Opin Genet Dev. 2012 Oct;22(5):416-22. doi: 10.1016/j.gde.2012.07.001. Epub 2012 Aug 3. Curr Opin Genet Dev. 2012. PMID: 22868173 Review.
Cited by
- Approaches for Benchmarking Single-Cell Gene Regulatory Network Methods.
Karamveer, Uzun Y. Karamveer, et al. Bioinform Biol Insights. 2024 Nov 4;18:11779322241287120. doi: 10.1177/11779322241287120. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39502448 Free PMC article. Review. - Functional Bidirectionality of ERV-Derived Long Non-Coding RNAs in Humans.
Song Y, Wen H, Zhai X, Jia L, Li L. Song Y, et al. Int J Mol Sci. 2024 Sep 29;25(19):10481. doi: 10.3390/ijms251910481. Int J Mol Sci. 2024. PMID: 39408810 Free PMC article. Review. - A suboptimal OCT4-SOX2 binding site facilitates the naïve-state specific function of a Klf4 enhancer.
Waite JB, Boytz R, Traeger AR, Lind TM, Lumbao-Conradson K, Torigoe SE. Waite JB, et al. PLoS One. 2024 Sep 30;19(9):e0311120. doi: 10.1371/journal.pone.0311120. eCollection 2024. PLoS One. 2024. PMID: 39348365 Free PMC article. - A dynamic regulome of shoot-apical-meristem-related homeobox transcription factors modulates plant architecture in maize.
Luo Z, Wu L, Miao X, Zhang S, Wei N, Zhao S, Shang X, Hu H, Xue J, Zhang T, Yang F, Xu S, Li L. Luo Z, et al. Genome Biol. 2024 Sep 19;25(1):245. doi: 10.1186/s13059-024-03391-8. Genome Biol. 2024. PMID: 39300560 Free PMC article.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials