Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors - PubMed (original) (raw)
Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors
Manuela Zoonens et al. Biophys J. 2008 Jul.
Abstract
The pH-dependent insertion of pHLIP across membranes is proving to be a useful property for targeting acidic tissues or tumors and delivering drugs attached to its C-terminus. It also serves as a model peptide for studies of protein insertion into membranes, so further elucidation of the insertion mechanism of pHLIP and its features is desirable. We examine how the peptide perturbs a model phosphatidylcholine membrane and how it associates with the lipid bilayer using an array of fluorescence techniques, including fluorescence anisotropy measurements of TMA-DPH anchored in bilayers, quenching of pHLIP fluorescence by brominated lipids and acrylamide, and measurements of energy transfer between aromatic residues of pHLIP and TMA-DPH. When pHLIP is bound to the surface of bilayers near neutral pH, the membrane integrity is preserved whereas the elastic properties of bilayers are changed as reported by an increase of membrane viscosity. When it is inserted, there is little perturbation of the lipids. The results also suggest that pHLIP can bind to the membrane surface in a shallow or a deep mode depending on the phase state of the lipids. Using parallax analysis, the change of the penetration depth of pHLIP was estimated to be 0.4 A from the bilayer center and 2.8 A from the membrane surface after the liquid-to-gel phase transition.
Figures
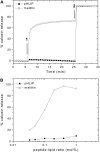
FIGURE 1
Effect of pHLIP versus melittin on the membrane leakage. (A) Kinetics of calcein leakage. The release of calcein (50 mM) encapsulated in POPC vesicles was monitored by the increase in fluorescence intensity at 515 nm (excitation at 450 nm), on addition of 0.2 mol % melittin or pHLIP (arrow 1). Complete leakage was achieved on addition of 0.05% Triton X-100 (arrow 2). The percentage of calcein release was calculated as described in Materials and Methods. The total lipid concentration was 0.1 mM and the buffer was 10 mM Mops, 150 mM NaCl, 5 mM EDTA, at pH 7.4. (B) Calcein release versus peptide concentrations. The intensity was recorded after 30 min incubation in the dark at room temperature.
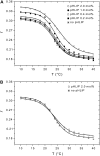
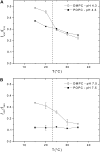
FIGURE 2
Effect of pHLIP on lipid membrane fluidity. The fluorescence anisotropy of TMA-DPH incorporated in DMPC bilayers was monitored in the presence of pHLIP at pH 7.5 (A) or pH 4.0 (B). Several concentrations of pHLIP (0.2, 0.4, 1.0, and 2.0 mol %) were tested with 1.9 mM DMPC LUVs containing 0.2 mol % TMA-DPH. The samples were excited by polarized light at 350 nm and emission was monitored at 420 nm. The fluorescence anisotropy was calculated as described in Materials and Methods and plotted as a function of temperature. Experimental points were fitted with a sigmoid function. The buffer was 20 mM Mops, at pH 7.5 or pH 4.0.
FIGURE 3
Quenched emission spectra of pHLIP by brominated lipids. pHLIP (0.25 mol %) was mixed with 0.6 mM unlabeled DMPC LUVs or DMPC LUVs containing 30 mol % (6,7)Br2-PC. Fluorescence emission spectra of pHLIP were recorded at 35°C (A and C) and 15°C (B and D). The buffer was 20 mM Mops, pH 7.5 (A and B) or pH 4.5 (C and D).
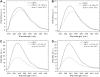
FIGURE 4
pH-dependence of FRET process between pHLIP and TMA-DPH incorporated in the membrane. pHLIP (0.33 mol %) was mixed with 0.85 mM POPC LUVs (A) or POPC LUVs containing 0.2 mol % TMA-DPH (B). The samples were excited at 280 nm at 20°C. The buffer was 20 mM Mops, at pH 7.5 or pH 4.3. Light scattering and fluorescence backgrounds of a blank containing TMA-DPH inserted into LUVs were subtracted from the spectra.
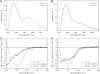
FIGURE 5
Temperature-dependence of pHLIP fluorescence in the presence of membranes. pHLIP (0.25 mol %) was mixed with either 0.85 mM POPC LUVs or DMPC LUVs at pH 4.3 (A) or pH 7.5 (B). Samples were excited at 280 nm and the emission fluorescence was observed as a function of temperature. Data are plotted as the ratio of intensities at 333 and 430 nm for samples at pH 4.3, and at 340 and 430 nm for samples at pH 7.5 to cancel out the effect of fluorescence quenching due to temperature variations. The dashed line indicates _T_m of phase transition for DMPC at 23°C.
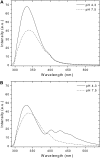
FIGURE 6
Insertion ability and secondary structure of wild-type and variant pHLIP peptides. The peptides (0.2 mol %) were separately mixed with 0.85 mM POPC LUVs containing 0.2 mol % TMA-DPH at pH 7.4 or pH 4.2. The emission fluorescence spectra of wild-type (A) and variant (B) peptides, after excitation at 280 nm, were corrected for light scattering and fluorescence background, and were normalized to the maximal intensity. The buffer was 20 mM Mops. The peptides (0.8 mol %) were analyzed by CD spectroscopy without or with 0.37 mM DMPC LUVs at 37°C, at pH 7.8 and pH 4.3. The CD spectra of wild-type (C) and variant (D) peptides were corrected for the background contribution, and were smoothed in a window of three points. The buffer was 2 mM Tris·HCl, 5 mM NaCl.
Similar articles
- Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane.
Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Reshetnyak YK, et al. Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15340-5. doi: 10.1073/pnas.0804746105. Epub 2008 Sep 30. Proc Natl Acad Sci U S A. 2008. PMID: 18829441 Free PMC article. - Comparison of lipid-dependent bilayer insertion of pHLIP and its P20G variant.
Vasquez-Montes V, Gerhart J, King KE, Thévenin D, Ladokhin AS. Vasquez-Montes V, et al. Biochim Biophys Acta Biomembr. 2018 Feb;1860(2):534-543. doi: 10.1016/j.bbamem.2017.11.006. Epub 2017 Nov 11. Biochim Biophys Acta Biomembr. 2018. PMID: 29138065 Free PMC article. - Bilayer Thickness and Curvature Influence Binding and Insertion of a pHLIP Peptide.
Karabadzhak AG, Weerakkody D, Deacon J, Andreev OA, Reshetnyak YK, Engelman DM. Karabadzhak AG, et al. Biophys J. 2018 May 8;114(9):2107-2115. doi: 10.1016/j.bpj.2018.03.036. Biophys J. 2018. PMID: 29742404 Free PMC article. - Biophysics of pH-Driven Membrane Insertion: A Review of the pHLIP Peptide.
Maia R, Ataka K, Heberle J, Baiz CR. Maia R, et al. J Phys Chem B. 2025 May 1;129(17):4123-4132. doi: 10.1021/acs.jpcb.5c00225. Epub 2025 Apr 18. J Phys Chem B. 2025. PMID: 40249795 Review. - pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents.
Andreev OA, Engelman DM, Reshetnyak YK. Andreev OA, et al. Mol Membr Biol. 2010 Oct;27(7):341-52. doi: 10.3109/09687688.2010.509285. Epub 2010 Oct 13. Mol Membr Biol. 2010. PMID: 20939768 Free PMC article. Review.
Cited by
- Membrane binding and insertion of a pHLIP peptide studied by all-atom molecular dynamics simulations.
Deng Y, Qian Z, Luo Y, Zhang Y, Mu Y, Wei G. Deng Y, et al. Int J Mol Sci. 2013 Jul 12;14(7):14532-49. doi: 10.3390/ijms140714532. Int J Mol Sci. 2013. PMID: 23857053 Free PMC article. - Cooperative Nonbonded Forces Control Membrane Binding of the pH-Low Insertion Peptide pHLIP.
Gupta C, Ren Y, Mertz B. Gupta C, et al. Biophys J. 2018 Dec 18;115(12):2403-2412. doi: 10.1016/j.bpj.2018.11.002. Epub 2018 Nov 7. Biophys J. 2018. PMID: 30503536 Free PMC article. - pHLIP Peptide Interaction with a Membrane Monitored by SAXS.
Narayanan T, Weerakkody D, Karabadzhak AG, Anderson M, Andreev OA, Reshetnyak YK. Narayanan T, et al. J Phys Chem B. 2016 Nov 10;120(44):11484-11491. doi: 10.1021/acs.jpcb.6b06643. Epub 2016 Oct 27. J Phys Chem B. 2016. PMID: 27726396 Free PMC article. - Heterogeneous diffusion of a membrane-bound pHLIP peptide.
Guo L, Gai F. Guo L, et al. Biophys J. 2010 Jun 16;98(12):2914-22. doi: 10.1016/j.bpj.2010.03.050. Biophys J. 2010. PMID: 20550904 Free PMC article. - Protonation Enhances the Inherent Helix-Forming Propensity of pHLIP.
Gupta C, Mertz B. Gupta C, et al. ACS Omega. 2017 Nov 30;2(11):8536-8542. doi: 10.1021/acsomega.7b01371. ACS Omega. 2017. PMID: 29214239 Free PMC article.
References
- Hunt, J. F., P. Rath, K. J. Rothschild, and D. M. Engelman. 1997. Spontaneous, pH-dependent membrane insertion of a transbilayer α-helix. Biochemistry. 36:15177–15192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources