Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases - PubMed (original) (raw)
. 2008 Apr 15;105(15):5809-14.
doi: 10.1073/pnas.0800940105. Epub 2008 Mar 21.
Edmond Chan, Pei-Qi Liu, Salvatore Orlando, Lin Zhang, Fyodor D Urnov, Michael C Holmes, Dmitry Guschin, Adam Waite, Jeffrey C Miller, Edward J Rebar, Philip D Gregory, Aaron Klug, Trevor N Collingwood
Affiliations
- PMID: 18359850
- PMCID: PMC2299223
- DOI: 10.1073/pnas.0800940105
Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases
Yolanda Santiago et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Gene knockout is the most powerful tool for determining gene function or permanently modifying the phenotypic characteristics of a cell. Existing methods for gene disruption are limited by their efficiency, time to completion, and/or the potential for confounding off-target effects. Here, we demonstrate a rapid single-step approach to targeted gene knockout in mammalian cells, using engineered zinc-finger nucleases (ZFNs). ZFNs can be designed to target a chosen locus with high specificity. Upon transient expression of these nucleases the target gene is first cleaved by the ZFNs and then repaired by a natural-but imperfect-DNA repair process, nonhomologous end joining. This often results in the generation of mutant (null) alleles. As proof of concept for this approach we designed ZFNs to target the dihydrofolate reductase (DHFR) gene in a Chinese hamster ovary (CHO) cell line. We observed biallelic gene disruption at frequencies >1%, thus obviating the need for selection markers. Three new genetically distinct DHFR(-/-) cell lines were generated. Each new line exhibited growth and functional properties consistent with the specific knockout of the DHFR gene. Importantly, target gene disruption is complete within 2-3 days of transient ZFN delivery, thus enabling the isolation of the resultant DHFR(-/-) cell lines within 1 month. These data demonstrate further the utility of ZFNs for rapid mammalian cell line engineering and establish a new method for gene knockout with application to reverse genetics, functional genomics, drug discovery, and therapeutic recombinant protein production.
Conflict of interest statement
Conflict of interest statement: Y.S., E.C., P.-Q.L., S.O., F.D.U., M.C.H, D.G., J.C.M., E.J.R., P.D.G., and T.N.C. are full-time employees of Sangamo BioSciences, Inc. A.K. is a member of the scientific advisory board for Sangamo BioSciences, Inc. L.Z. is a full-time employee of Pfizer, Inc.
Figures
Fig. 1.
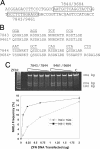
Targeting the DHFR locus with designed ZFNs. (A) Section of the DHFR gene targeted by ZFNs. The DNA sequence of the primary binding site for each ZFN is boxed. ZFN7843 and ZFN9461 bind the same 12-bp site. ZFN7844 binds the 12-bp site AATGCTCAGGTA, whereas ZFN9684 binds the 15-bp site AATGCTCAGGTACTG. (B) Recognition helix sequences of ZFNs. The sequence of the recognition helix from position −1 to +6 (27) is listed below its target triplet. Backbone sequences for the ZFPs can be found elsewhere (20). The C-terminal finger recognizes the 5′ most DNA triplet. The asterisks denote that these ZFNs employ the ELKK variant FokI domains throughout. (C) Comparison of ZFN activity. Plasmids encoding each pair of ZFNs (ZFN7843/ZFN7844 and ZFN9461/ZFN9684) containing the ELKK FokI variants were delivered in the amounts shown to CHO-S cells in suspension culture. The frequency of allelic mutation in each pool of treated cells was determined by using the CEL-I assay (gel). Bands migrating at 384, 204, and 180 bp represent the parent amplicon and the two CEL-I digestion products, respectively. The bands were quantitated by EtBr staining and densitometry to determine the frequency of NHEJ. The frequency of NHEJ is plotted against ZFN dosage. The lowest band on the 25-bp size ladder is 125 bp.
Fig. 2.
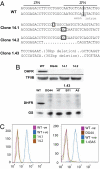
Genotype and phenotype of ZFN-induced _DHFR_−/− clones from CHO-S cells. (A) Each pair of sequences represents the two alleles of the DHFR gene in the designated cell line. For each mutant allele, inserted bases are boxed, and deleted bases are represented by dots. (B) Western blot for DHFR protein in WT CHO-S cells (WT) and the mutant cell lines 14/1, 14/2, and 1.43 and the commercially available DHFR-null CHO cell line DG44. Note that, for clone 1.43, we analyzed three independent subclones from this line (H7, B11, and A5). Input is normalized against TFIIB or glutamine synthetase (GS) expression levels as indicated. (C) Fluorescein-labeled methotrexate (FMTX)-based FACS analysis of _DHFR_−/− cell lines. WT(-ve), WT CHO-S cells not treated with FMTX; WT(+ve), WT CHO-S cells treated with FMTX. All mutant cell lines were treated with FMTX, including DG44 as a DHFR-null CHO cell control. (Left) Cells in adherent culture. (Right) Cells in suspension culture.
Fig. 3.
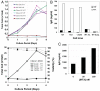
Growth and functional properties of the ZFN-generated _DHFR_−/− cell lines. (A) WT CHO-S (WT CHO) and ZFN-generated _DHFR_−/− CHO-S cell lines were cultured in the presence or absence of HT supplement as indicated. (Upper) Growth of _DHFR_−/− cell lines 14/1 and 14/2 compared with WT CHO cells in adherent culture conditions. (Lower) Growth and viability of the _DHFR_−/− cell line 1.43A5 in serum-free suspension culture. (B) IgG expression from WT or _DHFR_−/− cell lines after transfection with antibody expression constructs coexpressing a DHFR gene as a selection marker. Cells were cultured in the presence (+HT) or absence (−HT) of HT supplement for 14 days. IgG expression at >48 h was measured by ELISA. WT Ad., WT CHO-S cells in adherent medium; WT Susp., WT CHO-S cells in suspension medium. Cell lines 14/1 and 14/2 were cultured adherently. Cell line 1.43A5 was cultured in suspension. (C) Methotrexate selection/amplification in _DHFR_−/− cell line 14/1. Data shown is the level of IgG stably expressed from a clone 14/1 pool after 2 weeks of incubation in the absence of HT then 2 weeks more in the presence of the noted concentration of methotrexate (see Materials and Methods).
Comment in
- Knockout punches with a fistful of zinc fingers.
Wilson JH. Wilson JH. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5653-4. doi: 10.1073/pnas.0802298105. Epub 2008 Apr 9. Proc Natl Acad Sci U S A. 2008. PMID: 18401029 Free PMC article. Review. No abstract available.
Similar articles
- Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases.
Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, Collingwood TN, Gregory PD. Liu PQ, et al. Biotechnol Bioeng. 2010 May 1;106(1):97-105. doi: 10.1002/bit.22654. Biotechnol Bioeng. 2010. PMID: 20047187 - High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases.
Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, Joung JK, Voytas DF. Zhang F, et al. Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):12028-33. doi: 10.1073/pnas.0914991107. Epub 2010 May 27. Proc Natl Acad Sci U S A. 2010. PMID: 20508152 Free PMC article. - Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies.
Malphettes L, Freyvert Y, Chang J, Liu PQ, Chan E, Miller JC, Zhou Z, Nguyen T, Tsai C, Snowden AW, Collingwood TN, Gregory PD, Cost GJ. Malphettes L, et al. Biotechnol Bioeng. 2010 Aug 1;106(5):774-83. doi: 10.1002/bit.22751. Biotechnol Bioeng. 2010. PMID: 20564614 - Custom-designed zinc finger nucleases: what is next?
Wu J, Kandavelou K, Chandrasegaran S. Wu J, et al. Cell Mol Life Sci. 2007 Nov;64(22):2933-44. doi: 10.1007/s00018-007-7206-8. Cell Mol Life Sci. 2007. PMID: 17763826 Free PMC article. Review. - Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells.
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Durai S, et al. Nucleic Acids Res. 2005 Oct 26;33(18):5978-90. doi: 10.1093/nar/gki912. Print 2005. Nucleic Acids Res. 2005. PMID: 16251401 Free PMC article. Review.
Cited by
- Selection-independent generation of gene knockout mouse embryonic stem cells using zinc-finger nucleases.
Osiak A, Radecke F, Guhl E, Radecke S, Dannemann N, Lütge F, Glage S, Rudolph C, Cantz T, Schwarz K, Heilbronn R, Cathomen T. Osiak A, et al. PLoS One. 2011;6(12):e28911. doi: 10.1371/journal.pone.0028911. Epub 2011 Dec 14. PLoS One. 2011. PMID: 22194948 Free PMC article. - New routes for transgenesis of the mouse.
Belizário JE, Akamini P, Wolf P, Strauss B, Xavier-Neto J. Belizário JE, et al. J Appl Genet. 2012 Aug;53(3):295-315. doi: 10.1007/s13353-012-0096-y. Epub 2012 May 9. J Appl Genet. 2012. PMID: 22569888 Review. - Engineering lymphocyte subsets: tools, trials and tribulations.
June CH, Blazar BR, Riley JL. June CH, et al. Nat Rev Immunol. 2009 Oct;9(10):704-16. doi: 10.1038/nri2635. Nat Rev Immunol. 2009. PMID: 19859065 Free PMC article. Review. - Applications of Genome Editing Technologies in CAD Research and Therapy with a Focus on Atherosclerosis.
Mak MCE, Gurung R, Foo RSY. Mak MCE, et al. Int J Mol Sci. 2023 Sep 13;24(18):14057. doi: 10.3390/ijms241814057. Int J Mol Sci. 2023. PMID: 37762360 Free PMC article. Review. - Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells.
Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M. Inami Y, et al. J Cell Biol. 2011 Apr 18;193(2):275-84. doi: 10.1083/jcb.201102031. Epub 2011 Apr 11. J Cell Biol. 2011. PMID: 21482715 Free PMC article.
References
- Urlaub G, Kas E, Carothers AM, Chasin LA. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983;33:405–412. - PubMed
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. - PubMed
- Yamane-Ohnuki N, et al. Establishment of FUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng. 2004;87:614–622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials