Induced-fit or preexisting equilibrium dynamics? Lessons from protein crystallography and MD simulations on acetylcholinesterase and implications for structure-based drug design - PubMed (original) (raw)
Induced-fit or preexisting equilibrium dynamics? Lessons from protein crystallography and MD simulations on acetylcholinesterase and implications for structure-based drug design
Yechun Xu et al. Protein Sci. 2008 Apr.
Abstract
Crystal structures of acetylcholinesterase complexed with ligands are compared with side-chain conformations accessed by native acetylcholinesterase in molecular dynamics (MD) simulations. Several crystallographic conformations of a key residue in a specific binding site are accessed in a simulation of native acetylcholinesterase, although not seen in rotomer plots. Conformational changes upon ligand binding thus involve preexisting equilibrium dynamics. Consequently, rational drug design could benefit significantly from conformations monitored by MD simulations of native targets.
Figures
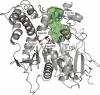
Figure 1.
3D structure of native _Tc_AChE (PDB access code 1ea5). Catalytic-triad residues are in red, the active-site gorge in green, and PAS Trp279 and CAS Trp84 in blue.
Figure 2.
χ1 and χ2 angles of Trp279 side-chain conformations from simulation, X-ray crystallography, and a rotomer library. (A) Conformations from a 20-ns MD simulation (gray dots) and 89 crystallographic AChE structures deposited in the PDB (black triangles). Simulated conformations form five islands. The white pentacle indicates the conformation in the crystal structure (1ea5) that the simulation is based on. Experimental conformations fall in seven groups: a, most native and complex structures; b, native _Dm_AChE (Harel et al. 2000) (1QO9); c, Tc_AChE/tacrine (Harel et al. 1993) (1ACJ); d, mAChE/TZ2PA6_syn (Bourne et al. 2004) (1Q83); e, mAChE/HI-6 (Ekstrom et al. 2006) (2GYU), mAChE/HLO-7 (2JEY), and mAChE/HLO-7/tabun (monomer A, 2JEZ) complexes; f, _Tc_AChE/A7 (Rydberg et al. 2006) (2CKM), _Tc_AChE/A8 (Rydberg et al. 2006) (1ODC), _Tc_AChE/NF595 (Colletier et al. 2006) (2CEK), mAChE/obidoxime (Ekstrom et al. 2006) (2GYW), and mAChE/HLO-7/tabun (monomer B, 2JEZ) complexes; g, mAChE/ortho-7 (Ekstrom et al. 2006) (2GYV) and mAChE/ortho-7/tabun (2JF0) complexes. (B) Most favorable conformations predicted by PROCHECK (Laskowski et al. 1993) based on an experimental library of rotomers (gray areas). The same experimental conformations as in A are presented (black triangles and white pentacle).
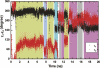
Figure 3.
Time dependence of χ1 and χ2 along the 20-ns trajectory. Colored bars indicate the five islands shown in Figure 2A. Khaki: island containing crystallographic groups a and b; blue: island containing group c; cyan: island containing group d; gray: island containing group f; mauve: island containing group g.
Similar articles
- Flexibility of aromatic residues in the active-site gorge of acetylcholinesterase: X-ray versus molecular dynamics.
Xu Y, Colletier JP, Weik M, Jiang H, Moult J, Silman I, Sussman JL. Xu Y, et al. Biophys J. 2008 Sep;95(5):2500-11. doi: 10.1529/biophysj.108.129601. Epub 2008 May 23. Biophys J. 2008. PMID: 18502801 Free PMC article. - Are induced fit protein conformational changes caused by ligand-binding predictable? A molecular dynamics investigation.
Gao C, Desaphy J, Vieth M. Gao C, et al. J Comput Chem. 2017 Jun 5;38(15):1229-1237. doi: 10.1002/jcc.24714. Epub 2017 Apr 16. J Comput Chem. 2017. PMID: 28419481 - Conformational flexibility in the peripheral site of Torpedo californica acetylcholinesterase revealed by the complex structure with a bifunctional inhibitor.
Colletier JP, Sanson B, Nachon F, Gabellieri E, Fattorusso C, Campiani G, Weik M. Colletier JP, et al. J Am Chem Soc. 2006 Apr 12;128(14):4526-7. doi: 10.1021/ja058683b. J Am Chem Soc. 2006. PMID: 16594661 - [Exploring the conformational energy landscape of acetylcholinesterase by kinetic crystallography].
Colletier JP, Weik M. Colletier JP, et al. Ann Pharm Fr. 2007 Mar;65(2):108-18. doi: 10.1016/s0003-4509(07)90024-3. Ann Pharm Fr. 2007. PMID: 17404544 Review. French. - Recent developments in structural studies on acetylcholinesterase.
Silman I, Sussman JL. Silman I, et al. J Neurochem. 2017 Aug;142 Suppl 2:19-25. doi: 10.1111/jnc.13992. Epub 2017 May 15. J Neurochem. 2017. PMID: 28503857 Review.
Cited by
- Chalcone Scaffolds Exhibiting Acetylcholinesterase Enzyme Inhibition: Mechanistic and Computational Investigations.
Malik YA, Awad TA, Abdalla M, Yagi S, Alhazmi HA, Ahsan W, Albratty M, Najmi A, Muhammad S, Khalid A. Malik YA, et al. Molecules. 2022 May 16;27(10):3181. doi: 10.3390/molecules27103181. Molecules. 2022. PMID: 35630658 Free PMC article. - Improving structure-based function prediction using molecular dynamics.
Glazer DS, Radmer RJ, Altman RB. Glazer DS, et al. Structure. 2009 Jul 15;17(7):919-29. doi: 10.1016/j.str.2009.05.010. Structure. 2009. PMID: 19604472 Free PMC article. - Acetylcholinesterase: from 3D structure to function.
Dvir H, Silman I, Harel M, Rosenberry TL, Sussman JL. Dvir H, et al. Chem Biol Interact. 2010 Sep 6;187(1-3):10-22. doi: 10.1016/j.cbi.2010.01.042. Epub 2010 Feb 4. Chem Biol Interact. 2010. PMID: 20138030 Free PMC article. - Consistent improvement of cross-docking results using binding site ensembles generated with elastic network normal modes.
Rueda M, Bottegoni G, Abagyan R. Rueda M, et al. J Chem Inf Model. 2009 Mar;49(3):716-25. doi: 10.1021/ci8003732. J Chem Inf Model. 2009. PMID: 19434904 Free PMC article. - Impact of Sucrose as Osmolyte on Molecular Dynamics of Mouse Acetylcholinesterase.
Lushchekina SV, Inidjel G, Martinez N, Masson P, Trovaslet-Leroy M, Nachon F, Koza MM, Seydel T, Peters J. Lushchekina SV, et al. Biomolecules. 2020 Dec 12;10(12):1664. doi: 10.3390/biom10121664. Biomolecules. 2020. PMID: 33322722 Free PMC article.
References
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., Hermans, J. Interaction models for water in relation to proteins hydration. In: Pullman B., editor. Intermolecular forces. Reidel; Dordrecht: 1981. pp. 331–342.
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A., Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690.
- Berendsen, H.J.C., van der Spoel, D., van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56.
- Colletier, J.P., Sanson, B., Nachon, F., Gabellieri, E., Fattorusso, C., Campiani, G., Weik, M. Conformational flexibility in the peripheral site of Torpedo californica acetylcholinesterase revealed by the complex structure with a bifunctional inhibitor. J. Am. Chem. Soc. 2006;128:4526–4527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources