RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes - PubMed (original) (raw)
RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes
David E Nowak et al. Mol Cell Biol. 2008 Jun.
Abstract
NF-kappaB plays a central role in cytokine-inducible inflammatory gene expression. Previously we empirically determined the identity of 92 members of the genetic network under direct NF-kappaB/RelA control that show marked heterogeneity in magnitude of transcriptional induction and kinetics of peak activation. To investigate this network further, we have applied a recently developed two-step chromatin immunoprecipitation assay that accurately reflects association and disassociation of RelA binding to its chromatin targets. Although inducible RelA binding occurs with similar kinetics on all NF-kappaB-dependent genes, serine 276 (Ser(276))-phosphorylated RelA binding is seen primarily on a subset of genes that are rapidly induced by tumor necrosis factor (TNF), including Gro-beta, interleukin-8 (IL-8), and IkappaBalpha. Previous work has shown that TNF-inducible RelA Ser(276) phosphorylation is controlled by a reactive oxygen species (ROS)-protein kinase A signaling pathway. To further understand the role of phospho-Ser(276) RelA in target gene expression, we inhibited its formation by ROS scavengers and antioxidants, treatments that disrupt phospho-Ser(276) formation but not the translocation and DNA binding of nonphosphorylated RelA. Here we find that phospho-Ser(276) RelA is required only for activation of IL-8 and Gro-beta, with IkappaBalpha being unaffected. These data were confirmed in experiments using RelA(-/-) murine embryonic fibroblasts reconstituted with a RelA Ser(276)Ala mutation. In addition, we observe that phospho-Ser(276) RelA binds the positive transcription elongation factor b (P-TEFb), a complex containing the cyclin-dependent kinase 9 (CDK-9) and cyclin T1 subunits. Inhibition of P-TEFb activity by short interfering RNA (siRNA)-mediated knockdown shows that the phospho-Ser(276) RelA-P-TEFb complex is required for IL-8 and Gro-beta gene activation but not for IkappaBalpha gene activation. These studies indicate that TNF induces target gene expression by heterogeneous mechanisms. One is mediated by phospho-Ser(276) RelA formation and chromatin targeting of P-TEFb controlling polymerase II (Pol II) recruitment and carboxy-terminal domain phosphorylation on the IL-8 and Gro-beta genes. The second involves a phospho-Ser(276) RelA-independent activation of genes preloaded with Pol II, exemplified by the IkappaBalpha gene. Together, these data suggest that the binding kinetics, selection of genomic targets, and mechanisms of promoter induction by RelA are controlled by a phosphorylation code influencing its interactions with coactivators and transcriptional elongation factors.
Figures
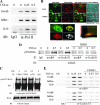
FIG. 1.
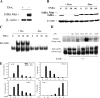
Dox-regulated NF-κB-dependent gene expression. (A) Tet-regulated IκBα Mut expression. HeLa Tet-Off cells expressing FLAG epitope-tagged IκBα Mut (Ser32Ala/Ser36Ala) were cultured in the presence (+) or absence (−) of 2 μg/ml Dox. Cytoplasmic extracts were extracted and assayed by Western blotting. (Top) Anti-FLAG Ab; (bottom) β-actin used as a loading control. (B) IκBα Mut is TNF resistant. Tet-regulated FLAG-IκBα Mut cells cultured in the presence or absence of Dox were TNF stimulated for the indicated times (min). (Top) Cytoplasmic extracts were assayed for IκBα abundance using anti-IκBα Ab. The endogenous and IκBα Mut isoforms are shown. (Bottom) β-Actin staining. (C) NF-κB binding in EMSA. Nuclear extracts from the experiment shown in panel B were assayed for NF-κB binding using EMSA. Shown are the bound complexes. The RelA/NF-κB1 (p50)-containing complex is indicated. (D) Supershift assay in EMSA. EMSA was performed on control (−) or TNF-stimulated (+) nuclear extracts in the absence or presence of indicated Ab (top). (Top) Long exposure (exp) of the supershifted (SShift) bands (indicated by *); (bottom) short exposure to demonstrate depletion of the TNF-inducible complex. The TNF-inducible NF-κB complex is composed predominantly of RelA·p50 heterodimers. (E) mRNA expression is NF-κB translocation dependent. RNA from a time series of TNF-stimulated HeLa IκBα Mut cells was analyzed by Q-RT-PCR. (The IκBα primers were designed to hybridize selectively to the 3′ untranslated region of the endogenous gene and do not detect IκBα Mut expression.) Shown are mRNA expression profiles in the presence or absence of Dox. For each gene, change is determined relative to unstimulated values.
FIG. 2.
Recruitment kinetics of NF-κB/RelA. (A) Specificity of ChIP assay. Two-step ChIP assay was performed on a time series of TNF-stimulated cells (39). Chromatin was subjected to IP using anti-RelA Ab. Eluted DNA was subjected to PCR using primers spanning the high-affinity RelA site located at nucleotides −99 to −61 in the human IL-8 promoter [IL-8 (Prox)] (top), primers to an upstream region of IL-8 not known to bind RelA and spanning nucleotides −1042 to −826 [(IL-8(Dist)] (middle), and primers spanning the Pol III-driven gene, U6 snRNA (bottom). Input DNA was used as a loading control. Shown is an ethidium bromide-stained agarose gel of the PCR products. Note that only the IL-8 (Prox) primers produce a TNF-inducible signal. (B) EMSA of TNF-stimulated cells. HeLa cells were exposed to either a 15-min pulse or continuous (tonic) stimulation. Nuclear extracts were subjected to EMSA using a high-affinity RelA-NF-κB1 binding site (10). Shown is an autoradiograph of the specifically bound species. (Top) Pulse stimulation; (bottom) tonic stimulation. (C) ChIP of tonic versus pulse TNF stimulation protocols. HeLa cells were stimulated for the indicated times after 15-min pulse (top) or tonic (bottom) TNF stimulation. Chromatin was prepared using two-step ChIP and immunoprecipitated with anti-RelA Ab. Shown is an ethidium bromide-stained gel of the PCR products. Input DNA was used as an IP control. (D) Time course of RelA recruitment to target genes. Cells were stained for indicated times and chromatin immunoprecipitated with anti-RelA Ab. Shown is an ethidium bromide-stained gel for PCR of the IL-8, IκBα, Gro-β, Naf1, NF-κB2, and TRAF1 genes by use of gene-specific primers flanking a high-affinity NF-κB binding site (56). Each PCR was run with 25 ng of genomic DNA as a positive control (PC). The negative control (NC) for the IP was IgG.
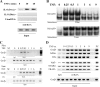
FIG. 3.
(A) Time course of phospho-Ser276 RelA formation. WCEs prepared from a time course of TNF-stimulated cells were fractionated by SDS-PAGE and blotted with anti-phospho-Ser276 RelA (top), anti-RelA (middle), and β-actin as an internal control (bottom). (B) Nucleus-cytoplasm distribution of phospho-Ser276 RelA. HeLa cells were stimulated with TNF for the times indicated at the top and fractionated into cytoplasmic and nuclear fractions. (Top) Western immunoblotting using anti-phospho-Ser276 RelA Ab. (Middle) Blot was reprobed for RelA as a loading control. Note the inducible accumulation of RelA in the nuclear fraction within 15 min of stimulation. (Bottom) β Lamin staining as a nuclear fraction control. The nuclear fractions stain strongly for β lamin. (C) Time course of phospho-Ser276 RelA recruitment to NF-κB-dependent genes. HeLa cells were subjected to a time course of TNF stimulation and chromatin was prepared. Shown is an ethidium bromide-stained gel of the PCR products. IgG, negative control for IP using nonimmune IgG. Each row represents a separate PCR assay. (D) Time course of phospho-Ser276 RelA binding in A549 cells. Experiment is as described for panel C. A549 cells show similar patterns of phospho-Ser276 RelA recruitment.
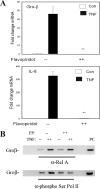
FIG. 4.
Inhibition of phospho-Ser276 RelA formation by antioxidant. (A) Kinetics of TNF-induced ROS formation and inhibition by DMSO. HeLa cells were loaded with DCFDA and stimulated with TNF (20 ng/ml) in the absence or presence of DMSO (0.5%, 2%). Fluorescence measurements were performed using a fluorescence-activated cell sorter at indicated times. Symbols: ▪, TNF; ▾, TNF plus 2% DMSO; ▴, TNF plus 0.5% DMSO; •, untreated. TNF induces detectable ROS formation within 15 min of stimulation. (Inset) Histograms of fluorescence-activated cell sorter analysis at 0, 30, and 60 min of treatment. (B) Kinetics of TNF-induced protein carbonylation. HeLa cells were stimulated with TNF for the times indicated. Shown is a Western blot for carbonylated proteins. Inducible carbonylation is seen within 15 min of stimulation (indicated by arrow). (C) Selective inhibition of TNF-induced phospho-Ser276 RelA formation by DMSO. HeLa cells were stimulated with TNF for 30 min the absence or presence of nontoxic concentrations of DMSO (2%) (26, 58). (Top) WCEs were prepared and blotted with anti-phospho-Ser276 RelA or pan anti-RelA Abs; (bottom) WCEs were blotted with anti-phospho-Ser536 RelA or anti-RelA Abs. (D) Antioxidant does not affect total RelA recruitment to target promoters. HeLa cells were stimulated for indicated times in the absence or presence of DMSO. Shown is a two-step ChIP assay using pan-anti-RelA Ab in the IP. Input is the loading control for the IP. DMSO has no impact on inducible RelA binding. (E) Effect of DMSO on NF-κB-dependent gene expression. mRNA was prepared from TNF-stimulated HeLa cells in the absence or presence of DMSO. Q-RT-PCR analysis was performed for the indicated genes. Shown is change relative to level at time zero and normalized to 18S RNA as an internal control. **, P < 0.01, t test. (F) Inhibition of TNF-induced phospho-Ser276 RelA formation by NAC. HeLa cells were stimulated with TNF for 30 min in the absence or presence of 10 mM NAC (26, 58). WCEs were prepared and blotted with anti-phospho-Ser276 RelA or anti-RelA Abs. (G) Effect of NAC on NF-κB-dependent gene expression. mRNA was prepared from a time course of TNF-stimulated HeLa cells in the absence or presence of NAC (10 mM). Q-RT-PCR analysis was performed as described for panel E. *, P < 0.05; **, P < 0.01, t test. (H) Antioxidant blocks phospho-Ser276 RelA recruitment. HeLa cells were TNF stimulated for the indicated times in the absence or presence of DMSO. Shown is a two-step ChIP assay using anti-phospho-Ser276 RelA Ab as the immunoprecipitating Ab. DMSO completely blocks TNF-inducible phospho-Ser276 RelA binding.
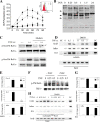
FIG. 5.
Effect of RelA Ser276Ala mutation on target gene expression. RelA−/− MEFs were transfected with expression vectors encoding a FLAG epitope-tagged monomeric strawberry fusion with RelAWT or RelA Ser276Ala, and pooled transfectants were isolated. Cells were stimulated for indicated times with TNF, and mRNA was extracted. Q-RT-PCR was performed to determine the activation of endogenous NF-κB-dependent genes.
FIG. 6.
TNF-inducible Pol II recruitment and RelA-P-TEFb complex formation. (A) Effect of TNF on Pol II loading. ChIP from TNF-stimulated HeLa cells, which were immunoprecipitated with anti-Pol II Ab. (B) Colocalization analysis. HeLa cells stably expressing EGFP-RelA were stimulated in the absence (control [Con]) or presence of TNF (30 min). Cells were fixed and stained with rabbit anti-CDK-9 Ab and Alexa Fluor 568-labeled goat anti-rabbit IgG. Nuclei were counterstained with TO-PRO-3 and are shown as overlays with differential interference contrast. (Bottom left) Sequential stages in image analysis with masking and colocalized pixels; (bottom right) corresponding two-dimensional histogram of EGFP-RelA and CDK-9 correlation. Shown is least-squares correlation analysis and the extent of colocalization for this image. After analysis of 39 individual cells, the Pearson's correlation coefficient was 0.428 ± 0.127, and the association was highly statistically significant (P < 0.001). (C) CDK-9 recruitment to RelA. FLAG-RelA-expressing HeLa cells were stimulated with TNF for 0 and 0.5 h in the absence or presence of DMSO as indicated. RelA-associated complexes were immunoprecipitated using anti-FLAG Ab, and the association of CDK-9 was detected by immunoblotting. (Bottom) IB of FLAG-RelA as a recovery control for the immunoprecipitation (IP). CDK-9-RelA complex association is TNF inducible and is reduced by DMSO treatment. (D) Coimmunoprecipitation assay for RelA-P-TEFb complex formation. HeLa cells were stimulated with TNF for 0 and 0.5 h in the absence or presence of antioxidant (DMSO). Nuclear extracts were prepared and immunoprecipitated with indicated anti-CDK-9 or Ccn T1 Abs. The presence of RelA in the immune complexes was then detected using anti-RelA Ab in the immunoblot (IB). A TNF-inducible complex was detected between Ser276-phosphorylated RelA, CDK-9, and Ccn T1. (Right) The inducible RelA-P-TEFb complex is quantitatively prevented by DMSO treatment. (E) Inducible P-TEFb binding. HeLa cells were stimulated with TNF for 0, 0.25, and 0.5 h in the absence or presence of DMSO as indicated. (Top) Two-step ChIP assay using anti-CDK-9 Ab as the immunoprecipitating Ab; (bottom) anti-Ccn T1 Ab was used. Shown is an ethidium bromide-stained gel of PCRs for IL-8, Gro-β, and IκBα as indicated (left). TNF-inducible recruitment of each member of the pTEF complex is rapid and indistinguishable. Inducible P-TEFb recruitment is significantly inhibited by DMSO.
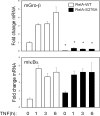
FIG. 7.
CDK inhibitor FP inhibits NF-κB-dependent gene expression by disrupting Pol II phospho-Ser2 CTD formation. (A) Gene expression HeLa cells were pretreated with vehicle or FP (500 nM) for 1 h prior to 30 min of TNF stimulation. mRNA expression was measured by Q-RT-PCR. Shown is change in mRNA normalized by 18S relative to unstimulated values. (B) FP inhibits P-TEFb-induced phospho-Ser Pol II CTD formation. HeLa cells were pretreated with vehicle or FP (500 nM) for 1 h prior to 30 min of TNF stimulation. (Top) Two-step ChIP assay using anti-RelA Ab as the immunoprecipitating Ab; (bottom) anti-phospho-Ser Pol II Ab was used. Although FP has no effect on inducible RelA binding, it significantly inhibits phospho-Ser Pol II recruitment.
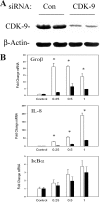
FIG. 8.
Effect of siRNA-mediated knockdown of CDK-9 on NF-κB-dependent gene expression. (A) Magnitude of siRNA-mediated knockdown. Cells were transfected with control (Con) or anti-CDK-9 siRNA prior to lysis and analysis of CDK-9 expression by Western blotting. Duplicate plates are shown for reproducibility. (Top) Staining with anti-CDK-9 Ab; the specific 42-kDa band is shown. (Bottom) Blot was reprobed with anti-β-actin Ab as a loading control. When normalized to β-actin, CDK-9 abundance is downregulated by ∼50%. (B) Effect of CDK-9 knockdown on inducible gene expression. Cells were transfected with control or CDK-9-targeted siRNA for 72 h prior to TNF stimulation for the indicated times. mRNA expression of early genes was measured by Q-RT-PCR. Shown is change in mRNA normalized by 18S relative to unstimulated values. *, P < 0.01, two-tailed t test.
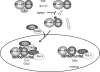
FIG. 9.
Model of NF-κB-dependent initiation of transcription. This schematic represents the steps involved in the two mechanistically distinct pathways involved in the transcriptional initiation of NF-κB-dependent gene expression mediated by inducible Pol II loading. In the cytoplasm, TNF stimulation releases RelA from sequestered cytoplasmic sites by inducing IκBα proteolysis. A delayed ROS signal mediates phospho-Ser276 RelA formation on a subset of proteins. Phospho-Ser276 RelA binds the CDK-9-Ccn T1 P-TEFb complex, recruiting it to IL-8 and Gro-β gene promoters. Here, P-TEFb is involved in phosphorylation of Ser2 in the Pol II CTD, NELF, and DSIF, resulting in productive transcriptional elongation. Transcriptional activation of the IL-8 and Gro-β genes is sensitive to antioxidant DMSO by its ability to disrupt RelA phosphorylation at Ser276 and thereby disrupt association with P-TEFb complexes. Transcriptional activation of the IL-8 and Gro-β genes is disrupted by FP and CDK-9 siRNA by blocking the serine phosphorylation of RNA Pol II CTD. Although phospho-Ser276 RelA binds IκBα, it is not functionally required for gene expression.
Similar articles
- TNF-alpha-induced NF-kappaB/RelA Ser(276) phosphorylation and enhanceosome formation is mediated by an ROS-dependent PKAc pathway.
Jamaluddin M, Wang S, Boldogh I, Tian B, Brasier AR. Jamaluddin M, et al. Cell Signal. 2007 Jul;19(7):1419-33. doi: 10.1016/j.cellsig.2007.01.020. Epub 2007 Jan 25. Cell Signal. 2007. PMID: 17317104 - ATM regulates NF-κB-dependent immediate-early genes via RelA Ser 276 phosphorylation coupled to CDK9 promoter recruitment.
Fang L, Choudhary S, Zhao Y, Edeh CB, Yang C, Boldogh I, Brasier AR. Fang L, et al. Nucleic Acids Res. 2014 Jul;42(13):8416-32. doi: 10.1093/nar/gku529. Epub 2014 Jun 23. Nucleic Acids Res. 2014. PMID: 24957606 Free PMC article. - RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection.
Brasier AR, Tian B, Jamaluddin M, Kalita MK, Garofalo RP, Lu M. Brasier AR, et al. J Virol. 2011 Nov;85(22):11752-69. doi: 10.1128/JVI.05360-11. Epub 2011 Sep 7. J Virol. 2011. PMID: 21900162 Free PMC article. - Expanding role of cyclin dependent kinases in cytokine inducible gene expression.
Brasier AR. Brasier AR. Cell Cycle. 2008 Sep 1;7(17):2661-6. doi: 10.4161/cc.7.17.6594. Epub 2008 Sep 12. Cell Cycle. 2008. PMID: 18728388 Free PMC article. Review. - RNA-driven cyclin-dependent kinase regulation: when CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners.
Michels AA, Bensaude O. Michels AA, et al. Biotechnol J. 2008 Aug;3(8):1022-32. doi: 10.1002/biot.200800104. Biotechnol J. 2008. PMID: 18655042 Review.
Cited by
- HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells.
Gringhuis SI, van der Vlist M, van den Berg LM, den Dunnen J, Litjens M, Geijtenbeek TB. Gringhuis SI, et al. Nat Immunol. 2010 May;11(5):419-26. doi: 10.1038/ni.1858. Epub 2010 Apr 4. Nat Immunol. 2010. PMID: 20364151 - Systems approaches to modeling chronic mucosal inflammation.
Kalita M, Tian B, Gao B, Choudhary S, Wood TG, Carmical JR, Boldogh I, Mitra S, Minna JD, Brasier AR. Kalita M, et al. Biomed Res Int. 2013;2013:505864. doi: 10.1155/2013/505864. Epub 2013 Oct 21. Biomed Res Int. 2013. PMID: 24228254 Free PMC article. - BRD4 Couples NF-κB/RelA with Airway Inflammation and the IRF-RIG-I Amplification Loop in Respiratory Syncytial Virus Infection.
Tian B, Yang J, Zhao Y, Ivanciuc T, Sun H, Garofalo RP, Brasier AR. Tian B, et al. J Virol. 2017 Feb 28;91(6):e00007-17. doi: 10.1128/JVI.00007-17. Print 2017 Mar 15. J Virol. 2017. PMID: 28077651 Free PMC article. - Coordinate activities of BRD4 and CDK9 in the transcriptional elongation complex are required for TGFβ-induced Nox4 expression and myofibroblast transdifferentiation.
Ijaz T, Jamaluddin M, Zhao Y, Zhang Y, Jay J, Finnerty CC, Herndon DN, Tilton RG, Brasier AR. Ijaz T, et al. Cell Death Dis. 2017 Feb 9;8(2):e2606. doi: 10.1038/cddis.2016.434. Cell Death Dis. 2017. PMID: 28182006 Free PMC article. - The potential for OGG1 inhibition to be a therapeutic strategy for pulmonary diseases.
Pan L, Boldogh I. Pan L, et al. Expert Opin Ther Targets. 2024 Mar;28(3):117-130. doi: 10.1080/14728222.2024.2317900. Epub 2024 Feb 14. Expert Opin Ther Targets. 2024. PMID: 38344773 Free PMC article. Review.
References
- Anrather, J., G. Racchumi, and C. Iadecola. 2005. cis-acting element-specific transcriptional activity of differentially phosphorylated nuclear factor-{kappa}B. J. Biol. Chem. 280244-252. - PubMed
- Babbar, N., and R. Casero, Jr. 2006. Tumor necrosis factor-A increases reactive oxygen species by inducing spermine oxidase in human lung epithelial cells: a potential mechanism for inflammation-induced carcinogenesis. Cancer Res. 6611125-11130. - PubMed
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 3327-337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI062885/AI/NIAID NIH HHS/United States
- P30 ES006676/ES/NIEHS NIH HHS/United States
- R01 AI040218/AI/NIAID NIH HHS/United States
- P30 ES06676/ES/NIEHS NIH HHS/United States
- R01 AI40218/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous