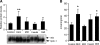Regulation of lipin-1 gene expression by glucocorticoids during adipogenesis - PubMed (original) (raw)
Regulation of lipin-1 gene expression by glucocorticoids during adipogenesis
Peixiang Zhang et al. J Lipid Res. 2008 Jul.
Abstract
Lipin-1 deficiency in the mouse causes generalized lipodystrophy, characterized by impaired adipose tissue development and insulin resistance. Lipin-1 expression in differentiating preadipocytes is required for normal expression of adipogenic transcription factors, including peroxisome proliferator-activated receptor gamma and CCAAT enhancer binding protein alpha, and for the synthesis of triacylglycerol. The requirement of lipin-1 for adipocyte differentiation can be explained, in part, by its activity as the sole adipocyte phosphatidic acid phosphatase-1 enzyme, which converts phosphatidate to diacylglycerol, the immediate precursor of triacylglycerol. Here we identify glucocorticoids as the stimulus for the induction of lipin-1 expression in differentiating adipocytes, and characterize a glucocorticoid response element (GRE) in the Lpin1 promoter. The Lpin1 GRE binds to the glucocorticoid receptor and leads to transcriptional activation in adipocytes and hepatocytes, as demonstrated by reporter gene transcription, electrophoretic mobility shift, and chromatin immunoprecipitation assays. This represents the first gene regulatory element identified to directly influence lipin-1 expression levels, and may modulate lipin-1 mRNA levels in adipose tissue and liver in conditions associated with increased local glucocorticoid concentrations in vivo, such as obesity and fasting.
Figures
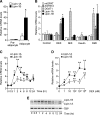
Fig. 1.
Glucocorticoid induces lipin-1 gene expression during adipocyte differentiation. A: Lipin-1 is dramatically induced in 3T3-L1 cells in response to adipocyte differentiation cocktail. Confluent 3T3-L1 cells (preadipocytes) were induced to differentiate with cocktail containing insulin, 3-isobutyl-
l
-methylxanthine (MIX), and dexamethasone (DEX) for 6 days (adipocytes). mRNA levels were determined by real-time RT-PCR and normalized to TATA box binding protein (TBP) mRNA levels. Values are expressed as the fold change compared with confluent cells (mean ± SD). B: Effects of differentiation cocktail components on expression of enzymes of triacylglycerol synthesis. Confluent 3T3-L1 preadipocytes were treated for 4 h with 10% FBS (control), DEX, MIX, insulin, or complete cocktail (DMI). mRNA levels for several enzymes in triacylglycerol biosynthesis were determined by real-time RT-PCR and expressed as fold difference compared with FBS-treated cells (mean ± SD). mtGPAT, mitochondrial glycerol-3-phosphate acyltransferase; AGPAT2, acylglycerolphosphate acyltransferase 2; DGAT1, diacylglycerol acyltransferase 1. C: Time course of DEX induction of lipin-1A and lipin-1B mRNA expression. Confluent 3T3-L1 preadipocytes were treated with DEX for the times indicated, and mRNA levels were expressed as fold difference compared with time 0 (mean ± SD). D: DEX induces lipin-1A and lipin-1B mRNA expression in a dose-dependent manner. Confluent 3T3-L1 preadipocytes were incubated with the indicated concentration of DEX in DMEM containing 10% FBS for 4 h, and mRNA levels were determined by real-time RT-PCR. E: Relative lipin-1A and -B isoform mRNA expression levels during DEX time course were determined by quantitative RT-PCR with primers that amplify both isoforms simultaneously. Products were resolved by agarose gel electrophoresis. Time of DEX treatment is indicated at bottom. TATA box binding protein was amplified as a control. For all studies in A–D, n = 3; * P < 0.05, ** P < 0.01 versus corresponding controls.
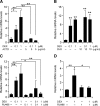
Fig. 2.
Lipin-1 regulation by glucocorticoid occurs at the transcriptional level. Results in panels A–C are for lipin-1B; the analogous experiments for lipin-1A are presented in supplementary Figure I, panels A–C. A: Induction of lipin-1B expression by DEX is inhibited by the RNA synthesis inhibitor actinomycin D (ACTD). Confluent 3T3-L1 preadipocytes were treated with the indicated concentration of DEX in the absence or presence of ACTD for 4 h. B: Induction of lipin-1B was not inhibited by the protein synthesis inhibitor cycloheximide (CHX). Confluent 3T3-L1 preadipocytes were treated with the indicated concentration of DEX in the absence or presence of CHX for 4 h. C: Induction of lipin-1B was inhibited by the glucocorticoid receptor (GR) antagonist RU486. Confluent 3T3-L1 preadipocytes were treated with the indicated concentration of DEX in the absence or presence of RU486 for 4 h. D: Human primary preadipocytes were treated with 1 μM DEX in the absence or presence of 1 μM RU486 for 4 h and exhibit the same response of lipin-1 gene expression as observed in 3T3-L1 cells. The analogous experiment in mature human primary adipocytes is presented in supplementary Figure ID. For all studies, values represent the mean ± SD for three samples. * P < 0.05, **P < 0.01 versus controls or for comparisons indicated.
Fig. 3.
Glucocorticoid-induced lipin-1 mRNA expression leads to increased lipin-1 protein and phosphatidic acid phosphatase-1 (PAP1) activity. 3T3-L1 adipocytes (differentiated for 8 days) were incubated in 1% FBS overnight, then treated for 8 h with DEX, MIX, insulin, or DMI mixture. A: Cell lysates were analyzed for lipin-1 protein levels by Western blotting with rabbit polyclonal anti-lipin-1 antibody and bands quantitated (n = 8). A representative gel is shown at bottom with duplicate samples from each treatment. Note that the lanes labeled DMI are from the same gel, but are shown as a separate box because of removal of irrelevant lanes in the figure. A nonspecific band is indicated (NS). B: N-ethylmaleimide-inhibitable PAP1 activity in cell lysates from samples in A. PAP1 activity was normalized to total cell protein. n = 6; * P < 0.05, ** P < 0.01 versus control treatment. Values presented represent mean ± SD.
Fig. 4.
Dexamethasone activation of the Lpin1 promoter is mediated by a glucocorticoid response element (GRE) motif. A: Transcriptional activity of Lpin1 promoter-luciferase constructs under basal conditions (10% FBS). 3T3-L1 preadipocytes were cotransfected with Lpin1 promoter luciferase constructs, renilla-luciferase control vector, and rat GR expression vector (pSG5-GR). Lpin1 promoter activity was normalized to renilla luciferase activity. Data represent the mean ± SE of four samples, expressed as the ratio of the Lpin1 promoter segment to that of the pGL3-basic vector. B: Effect of 1 μM DEX on the activity of the _Lpin1_-luciferase constructs in 3T3-L1 cells. Luciferase activity was measured 24 h after treatment with vehicle (DMSO) or DEX. Data are shown as fold difference (mean ± SE) between DEX and the vehicle-treated groups. n = 4; * P < 0.05 versus vehicle treated. C: Sequence of the Lpin1 promoter region containing the GRE (−311 to −297). The wild-type (wt) putative GRE motif and the mutant (mt) GRE (−311/−297) used in subsequent studies are in boldface. D: Effect of 1 μM DEX on Lpin1 promoter-luciferase constructs containing wild-type and mutant versions of the GRE. n = 4; * P < 0.05 versus vehicle-treated.
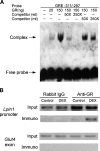
Fig. 5.
The lipin-1 GRE binds GR. A: Analysis of lipin-1 GRE binding to GR by electrophoretic mobility shift assay (EMSA). An EMSA was performed using purified human GR and a biotinylated oligonucleotide probe containing the lipin-1 GRE and surrounding sequences (−311 to −297; see Fig. 4C). Competition assays were performed by preincubating the reaction for 25 min with 50- or 250-fold molar excess of unlabeled oligonucleotides containing wild-type (wt) or mutant (mt) GRE sequences. Arrows indicate position of unbound oligonucleotide probe and probe complexed with GR. Results are representative of three independent experiments. B: Chromatin immunoprecipitation (ChIP) assay to detect the binding of GR to the Lpin1 promoter in chromatin from 3T3-L1 adipocytes. Cells were treated with vehicle (DMSO) or DEX for 4 h, and association of the GR with Lpin1 promoter sequences (−632/−169) was detected by immunoprecipitation with anti-GR antibody followed by PCR amplification of Lpin1 promoter sequences. Input lanes show presence of Lpin1 sequence before immunoprecipitation in all samples. Rabbit IgG was used as a negative control for specificity of the immunoprecipitation, and an exon of the Glut4 gene was used as a negative control for the PCR amplification.
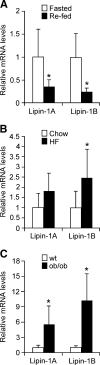
Fig. 6.
Adipose tissue lipin-1 expression is increased in conditions associated with increased local glucocorticoid levels_._ A: Lipin-1 mRNA levels in epididymal adipose tissue from C57BL/6J mice after a 16 h fast (fasted) or after a 16 h fast plus a 4 h re-feeding (re-fed). mRNA levels were determined by real-time RT-PCR and normalized to TBP mRNA levels. n = 6; * P < 0.05 versus fasted samples. B: Lipin-1 levels in epididymal adipose tissue from C57BL/6J mice fed a chow or high-fat diet for 16 weeks. n = 6; * P < 0.05 high-fat diet versus chow diet. C: Lipin-1 levels in epididymal adipose tissue of ob/ob and wild-type (wt) littermates. The value of wt was set to 1, and levels in ob/ob mice were expressed relative to wt. n = 5; * P < 0.05. Data are shown as mean ± SEM.
Similar articles
- Lipin-1 phosphatidic phosphatase activity modulates phosphatidate levels to promote peroxisome proliferator-activated receptor γ (PPARγ) gene expression during adipogenesis.
Zhang P, Takeuchi K, Csaki LS, Reue K. Zhang P, et al. J Biol Chem. 2012 Jan 27;287(5):3485-94. doi: 10.1074/jbc.M111.296681. Epub 2011 Dec 6. J Biol Chem. 2012. PMID: 22157014 Free PMC article. - Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro.
Phan J, Péterfy M, Reue K. Phan J, et al. J Biol Chem. 2004 Jul 9;279(28):29558-64. doi: 10.1074/jbc.M403506200. Epub 2004 Apr 29. J Biol Chem. 2004. PMID: 15123608 - Redundant roles of the phosphatidate phosphatase family in triacylglycerol synthesis in human adipocytes.
Temprano A, Sembongi H, Han GS, Sebastián D, Capellades J, Moreno C, Guardiola J, Wabitsch M, Richart C, Yanes O, Zorzano A, Carman GM, Siniossoglou S, Miranda M. Temprano A, et al. Diabetologia. 2016 Sep;59(9):1985-94. doi: 10.1007/s00125-016-4018-0. Epub 2016 Jun 25. Diabetologia. 2016. PMID: 27344312 Free PMC article. - Biphasic expression of lipin suggests dual roles in adipocyte development.
Phan J, Peterfy M, Reue K. Phan J, et al. Drug News Perspect. 2005 Jan-Feb;18(1):5-11. doi: 10.1358/dnp.2005.18.1.877165. Drug News Perspect. 2005. PMID: 15753971 Review. - The lipin protein family: dual roles in lipid biosynthesis and gene expression.
Reue K, Zhang P. Reue K, et al. FEBS Lett. 2008 Jan 9;582(1):90-6. doi: 10.1016/j.febslet.2007.11.014. Epub 2007 Nov 20. FEBS Lett. 2008. PMID: 18023282 Free PMC article. Review.
Cited by
- Dietary cholesterol reduces plasma triacylglycerol in apolipoprotein E-null mice: suppression of lipin-1 and -2 in the glycerol-3-phosphate pathway.
Obama T, Nagaoka S, Akagi K, Kato R, Horiuchi N, Horai Y, Aiuchi T, Arata S, Yamaguchi T, Watanabe M, Itabe H. Obama T, et al. PLoS One. 2011;6(8):e22917. doi: 10.1371/journal.pone.0022917. Epub 2011 Aug 9. PLoS One. 2011. PMID: 21857965 Free PMC article. - Forkhead box A3 mediates glucocorticoid receptor function in adipose tissue.
Ma X, Xu L, Mueller E. Ma X, et al. Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):3377-82. doi: 10.1073/pnas.1601281113. Epub 2016 Mar 8. Proc Natl Acad Sci U S A. 2016. PMID: 26957608 Free PMC article. - Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling.
Coleman RA, Mashek DG. Coleman RA, et al. Chem Rev. 2011 Oct 12;111(10):6359-86. doi: 10.1021/cr100404w. Epub 2011 Jun 1. Chem Rev. 2011. PMID: 21627334 Free PMC article. Review. No abstract available. - Cardiac lipin 1 expression is regulated by the peroxisome proliferator activated receptor γ coactivator 1α/estrogen related receptor axis.
Mitra MS, Schilling JD, Wang X, Jay PY, Huss JM, Su X, Finck BN. Mitra MS, et al. J Mol Cell Cardiol. 2011 Jul;51(1):120-8. doi: 10.1016/j.yjmcc.2011.04.009. Epub 2011 Apr 28. J Mol Cell Cardiol. 2011. PMID: 21549711 Free PMC article. - Silencing Mediator of Retinoid and Thyroid Hormone Receptors (SMRT) regulates glucocorticoid action in adipocytes.
Emont MP, Mantis S, Kahn JH, Landeche M, Han X, Sargis RM, Cohen RN. Emont MP, et al. Mol Cell Endocrinol. 2015 May 15;407:52-6. doi: 10.1016/j.mce.2015.03.002. Epub 2015 Mar 9. Mol Cell Endocrinol. 2015. PMID: 25766503 Free PMC article.
References
- Péterfy M., J. Phan, P. Xu, and K. Reue. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27 121–124. - PubMed
- Reue K., P. Xu, X. P. Wang, and B. G. Slavin. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41 1067–1076. - PubMed
- Donkor J., M. Sariahmetoglu, J. Dewald, D. N. Brindley, and K. Reue. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282 3450–3457. - PubMed
- Phan J., and K. Reue. 2005. Lipin, a lipodystrophy and obesity gene. Cell Metab. 1 73–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases