Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP - PubMed (original) (raw)
Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP
Guillaume M Hautbergue et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Adaptor proteins stimulate the nuclear export of mRNA, but their mechanism of action remains unclear. Here, we show that REF/ALY binds mRNA; but upon formation of a ternary complex with TAP the RNA is transferred from REF to TAP, and overexpression of TAP displaces REF from mRNA in vivo. RNA is also handed over from two other adaptors, 9G8 and SRp20 to TAP upon formation of a ternary complex. Interestingly, the RNA-binding affinity of TAP is enhanced 4-fold in vitro once it is complexed with REF. 9G8 and SRp20 also enhance the TAP RNA-binding activity in vitro. Consistent with a model in which TAP directly binds mRNA handed over from adaptors during export, we show that TAP binds mRNA in vivo by an arginine-rich motif in its N-terminal domain. The importance of direct TAP-mRNA interactions is confirmed by the observation that a mutant form of TAP that fails to bind mRNA but retains the ability to bind REF does not function in mRNA export.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
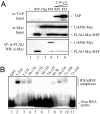
Displacement of UAP56 from REF by TAP. (A) Co-IP assays. Total extracts from 293T cells (Mock, lane 1) transfected with FLAG-Myc-tagged REF (lane 2) or cotransfected with Myc-tagged UAP56 and either control (FLAG) or FLAG-Myc-tagged REF construct (REF) (lanes 3–6) were incubated with increasing amount of purified recombinant GST-TAP-p15 (TAP-p15, lanes 5 and 6) before IP with α-FLAG antibodies. Total extracts (Top and Middle) and purified complexes (Bottom) were analyzed by Western blotting (WB) with the indicated antibodies. (B) EMSA with a 32P-radiolabeled 15-mer RNA in the presence of 1 μM GST control (lane 1) and GST-tagged fusions of REF (lanes 2–11).
Fig. 2.
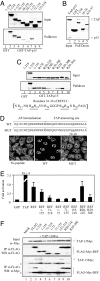
TAP-binding domain in REF. (A) Pulldown assays. GST (control, lane 1) and GST-TAP-p15 (lanes 2–9) were immobilized on glutathione-coated beads. 35S-radiolabeled REF proteins indicated by aa numbers were added to the binding reactions in the presence of RNase. Eluted proteins were analyzed by SDS/PAGE and PhosphorImaging. (B) Pulldown assays. Input corresponds to purified TAP-p15. Recombinant GST (lane 2) and GST-REF (lane 3), GST-REF amino acids 16–36 (lane 4), or GST-REF 1–155 deleted from amino acids 16 to 37 (lane 5) were immobilized on glutathione beads, and TAP-p15 was added. Eluted proteins were analyzed by SDS/PAGE stained with Coomassie blue. (C) Pulldown assays as in A. GST (lane 1) and GST-TAP-p15 (lanes 2–10) were used in pulldown assays with 35S-REF proteins + RNase. Sequence of the TAP-interacting site of REF with underlined arginine positions is shown. (D) (Upper) Peptide sequences of Antennapedia cell-permeable element (AP) fused to REF amino acids 16–36 (WT) or to the same peptide with mutations of R29,R30 (MUT). (Lower) Poly(A)+ RNA localization in HeLa cells (no peptide) and in cells incubated for 72 h with 12.5 μM WT or MUT peptides. (E) Luciferase activity generated by MS2 fusions normalized with a LacZ transfection control in the tethered export assay with the pLUCSALRRE6MS2 reporter. Error bars represent SD values from three datasets, each carried out in triplicate. Fold activations relative to MS2-GFP are shown. (F) Co-IP assays. Total extracts from 293T cells (Mock) cotransfected with Myc-tagged TAP and either control (FLAG) or FLAG-Myc-tagged REF constructs indicated by aa numbers were treated with RNase before IP with α-FLAG antibodies. Total extracts (Top) and purified complexes (Middle and Bottom) were analyzed by Western blotting with the indicated antibodies. Asterisks indicate heavy or light IgG chains. REF amino acids 1–155 and 71–218 comigrate with the light chains of FLAG antibody used for IP.
Fig. 3.
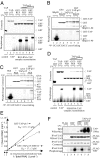
Export adaptors enhance TAP RNA binding. (A) In vitro reconstitution of REF-RNA-TAP-p15 complexes. GST (lane 1), TAP-p15 (lanes 2 and 3), or GST-REF (REF, lanes 4–9) were first incubated with continuously 32P-radiolabeled RNA. Equimolar (lanes 6 and 7) or 5 molar excess (lanes 8 and 9) TAP-p15 was added to the GST and GST-REF RNA-binding reactions. Bound RNA was UV-cross-linked as indicated, treated with RNase, and protein–RNA complexes were purified. Eluted complexes were analyzed on SDS/PAGE by Coomassie blue (Upper) and PhosphorImaging (Lower). (B and C) Cross-linking experiments carried out as described in A except GST-TAP was used to pull down SRp20 (amino acids 1–90) (B) or 9G8 (amino acids 12–98) (C). The indicated end-labeled RNA oligonucleotides were used. Lanes 1 and 2 show free SR proteins; lanes 3–8 represent pulldowns. (D) In vitro reconstitution of 9G8 RNA-TAP-p15 complexes. Experiments were carried out and displayed as in A except GST-9G8 (amino acids 1–122) was used. (E) RNA binding affinities were measured as described in Experimental Procedures for GST-TAP-p15 and GST-TAP-p15–REF2–1 complex. (F) In vivo competition assay. 293T cells transfected with either FLAG control (FLAG) or FLAG-tagged REF (REF) and increasing amounts of Myc-tagged TAP and p15 expression plasmids (TAP-p15) were exposed to UV as indicated. REF complexes were immunopurified with α-FLAG antibodies. (Bottom) Purified complexes were treated with RNase, and the resulting residual mRNAs cross-linked to REF were end-labeled by using [γ32]ATP and polynucleotide kinase before PhosphorImaging. (Top) Increasing expression of TAP was confirmed in total extracts by α-Myc Western blotting (WB). (Top Middle and Bottom Middle) Successful purification of FLAG-REF was confirmed by WB (Top Middle) and Coomassie staining (Bottom Middle). The band found in all lanes in Bottom Middle corresponds to the IgG light chain.
Fig. 4.
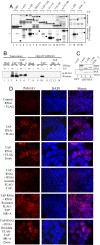
Identification of the TAP RNA-binding domain. (A) RNA cross-linking using continuously labeled RNA and GB1-TAP fusions. (B) UV cross-linking mRNP capture assay. 293T cells were transfected with FLAG-TAP-Myc or FLAG-TAP-Myc 10RA vectors and mRNA capture assays carried out after UV irradiation of cells as indicated. TAP proteins cross-linked to the mRNA were detected by Western blotting (WB) with α-Myc Abs. (C) GST pulldowns using GST (lane 1) or GST-REF (lanes 2–4) with GB1-TAP fusions. (D) Rescue of the mRNA export block induced by TAP RNAi. HeLa cells were transfected with pSUPERLUC (control RNAi) or pSUPERTAP (TAP RNAi) together with FLAG, FLAG-TAP, or FLAG-TAP 10RA vectors modified to make the mRNAs resistant to RNAi. Poly(A)+ RNA was detected by using Cy3-oligo(dT)50. The third and sixth sets of panels from the Top show a larger magnification image compared with other panels.
Similar articles
- Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1.
Hung ML, Hautbergue GM, Snijders AP, Dickman MJ, Wilson SA. Hung ML, et al. Nucleic Acids Res. 2010 Jun;38(10):3351-61. doi: 10.1093/nar/gkq033. Epub 2010 Feb 2. Nucleic Acids Res. 2010. PMID: 20129943 Free PMC article. - SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs.
Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. Escudero-Paunetto L, et al. Virology. 2010 Jun 5;401(2):155-64. doi: 10.1016/j.virol.2010.02.023. Epub 2010 Mar 12. Virology. 2010. PMID: 20227104 Free PMC article. - Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. Hargous Y, et al. EMBO J. 2006 Nov 1;25(21):5126-37. doi: 10.1038/sj.emboj.7601385. Epub 2006 Oct 12. EMBO J. 2006. PMID: 17036044 Free PMC article. - Structure-function relationships in the Nab2 polyadenosine-RNA binding Zn finger protein family.
Fasken MB, Corbett AH, Stewart M. Fasken MB, et al. Protein Sci. 2019 Mar;28(3):513-523. doi: 10.1002/pro.3565. Epub 2019 Jan 16. Protein Sci. 2019. PMID: 30578643 Free PMC article. Review. - mRNA export and cancer.
Siddiqui N, Borden KL. Siddiqui N, et al. Wiley Interdiscip Rev RNA. 2012 Jan-Feb;3(1):13-25. doi: 10.1002/wrna.101. Epub 2011 Jul 27. Wiley Interdiscip Rev RNA. 2012. PMID: 21796793 Review.
Cited by
- Comparative genomics of proteins involved in RNA nucleocytoplasmic export.
Serpeloni M, Vidal NM, Goldenberg S, Avila AR, Hoffmann FG. Serpeloni M, et al. BMC Evol Biol. 2011 Jan 11;11:7. doi: 10.1186/1471-2148-11-7. BMC Evol Biol. 2011. PMID: 21223572 Free PMC article. - Germline heterozygous mutations in Nxf1 perturb RNA metabolism and trigger thrombocytopenia and lymphopenia in mice.
Chappaz S, Law CW, Dowling MR, Carey KT, Lane RM, Ngo LH, Wickramasinghe VO, Smyth GK, Ritchie ME, Kile BT. Chappaz S, et al. Blood Adv. 2020 Apr 14;4(7):1270-1283. doi: 10.1182/bloodadvances.2019001323. Blood Adv. 2020. PMID: 32236527 Free PMC article. - Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome.
Fáberová V, Kalasová I, Krausová A, Hozák P. Fáberová V, et al. Cells. 2020 May 11;9(5):1191. doi: 10.3390/cells9051191. Cells. 2020. PMID: 32403279 Free PMC article. - The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1.
Uranishi H, Zolotukhin AS, Lindtner S, Warming S, Zhang GM, Bear J, Copeland NG, Jenkins NA, Pavlakis GN, Felber BK. Uranishi H, et al. J Biol Chem. 2009 Sep 18;284(38):26106-16. doi: 10.1074/jbc.M109.040113. Epub 2009 Jul 8. J Biol Chem. 2009. PMID: 19586903 Free PMC article. - Interferon-Regulated Expression of Cellular Splicing Factors Modulates Multiple Levels of HIV-1 Gene Expression and Replication.
Roesmann F, Müller L, Klaassen K, Heß S, Widera M. Roesmann F, et al. Viruses. 2024 Jun 11;16(6):938. doi: 10.3390/v16060938. Viruses. 2024. PMID: 38932230 Free PMC article. Review.
References
- Strasser K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. - PubMed
- Cheng H, et al. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/D010519/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F000588/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- GR077544A1A/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous