The role of disorder in interaction networks: a structural analysis - PubMed (original) (raw)
The role of disorder in interaction networks: a structural analysis
Philip M Kim et al. Mol Syst Biol. 2008.
Abstract
Recent studies have emphasized the value of including structural information into the topological analysis of protein networks. Here, we utilized structural information to investigate the role of intrinsic disorder in these networks. Hub proteins tend to be more disordered than other proteins (i.e. the proteome average); however, we find this only true for those with one or two binding interfaces ('single'-interface hubs). In contrast, the distribution of disordered residues in multi-interface hubs is indistinguishable from the overall proteome. Surprisingly, we find that the binding interfaces in single-interface hubs are highly structured, as is the case for multi-interface hubs. However, the binding partners of single-interface hubs tend to have a higher level of disorder than the proteome average, suggesting that their binding promiscuity is related to the disorder of their binding partners. In turn, the higher level of disorder of single-interface hubs can be partly explained by their tendency to bind to each other in a cascade. A good illustration of this trend can be found in signaling pathways and, more specifically, in kinase cascades. Finally, our findings have implications for the current controversy related to party and date-hubs.
Figures
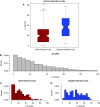
Figure 1
(A) Disorder in singlish-interface hubs versus multi-interface hubs. (Wilcoxon ranksum test, _P_=1.0e−8). (B) Distribution of disordered residues in the yeast proteins. As can be seen, most yeast proteins have a relatively low level of disorder; however, almost all have some fraction of disordered residues. (C) Distribution of disorder in multi-interface hubs. The distribution does not significantly deviate from the distribution of the yeast proteome (Kolmogorov-Smirnov test, _P_-value=0.11). In other words, multi-interface hubs do not have different levels of disorder than normal proteins. (D) Distribution of disorder in singlish-interface hubs. Singlish-interface hubs show a different distribution in terms of disorder (Kolmogorov–Smirnov test, _P_=2.0e−6).
Figure 2
dN/dS ratio of ordered and disordered proteins, and hubs. (A) All yeast proteins, split by order/disorder (Wilcoxon rank sum test, _P_-value <2.2e−16); (B) Hubs only (Wilcoxon rank sum test, _P_=1.5e−6).
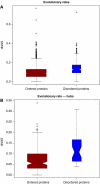
Figure 3
(A) Disorder in the interface regions of singlish- and multi-interface hubs. (Wilcoxon ranksum test, _P_=0.4). (B) Disorder of the binding partner of singlish- and multi-interface hubs (Wilcoxon ranksum test, _P_=4.5e−5). BIOGRID data are reported in the boxplot. Similar results were found for Kim et al (2006) and Batada et al (2006) data sets (Supplementary Figure S5). (C) Schematic representation of disorder in singlish-interface hubs. Singlish-interface hubs have large portions of disordered regions (painted in gray). However, the interface is itself is highly structured (painted in black). One reason for the disorder in the bulk of the protein is the fact that singlish-interface hubs often are targeted by kinases. On the other hand, they tend to be kinases themselves and target disordered regions in other proteins.
Similar articles
- What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae?
Ekman D, Light S, Björklund AK, Elofsson A. Ekman D, et al. Genome Biol. 2006;7(6):R45. doi: 10.1186/gb-2006-7-6-r45. Genome Biol. 2006. PMID: 16780599 Free PMC article. - Dynamic hubs show competitive and static hubs non-competitive regulation of their interaction partners.
Goel A, Wilkins MR. Goel A, et al. PLoS One. 2012;7(10):e48209. doi: 10.1371/journal.pone.0048209. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23118954 Free PMC article. - Predicting the binding patterns of hub proteins: a study using yeast protein interaction networks.
Andorf CM, Honavar V, Sen TZ. Andorf CM, et al. PLoS One. 2013;8(2):e56833. doi: 10.1371/journal.pone.0056833. Epub 2013 Feb 19. PLoS One. 2013. PMID: 23431393 Free PMC article. - Protein dynamics and conformational disorder in molecular recognition.
Mittag T, Kay LE, Forman-Kay JD. Mittag T, et al. J Mol Recognit. 2010 Mar-Apr;23(2):105-16. doi: 10.1002/jmr.961. J Mol Recognit. 2010. PMID: 19585546 Review. - What can we learn from highly connected beta-rich structures for structural interface design?
Emekli U, Gunasekaran K, Nussinov R, Haliloglu T. Emekli U, et al. Methods Mol Biol. 2008;474:235-53. doi: 10.1007/978-1-59745-480-3_12. Methods Mol Biol. 2008. PMID: 19031068 Review.
Cited by
- Intrinsic protein disorder reduces small-scale gene duplicability.
Banerjee S, Feyertag F, Alvarez-Ponce D. Banerjee S, et al. DNA Res. 2017 Aug 1;24(4):435-444. doi: 10.1093/dnares/dsx015. DNA Res. 2017. PMID: 28430886 Free PMC article. - Protein-Protein Interactions Mediated by Intrinsically Disordered Protein Regions Are Enriched in Missense Mutations.
Wong ETC, So V, Guron M, Kuechler ER, Malhis N, Bui JM, Gsponer J. Wong ETC, et al. Biomolecules. 2020 Jul 24;10(8):1097. doi: 10.3390/biom10081097. Biomolecules. 2020. PMID: 32722039 Free PMC article. - Intrinsic disorder and protein multibinding in domain, terminal, and linker regions.
Fong JH, Panchenko AR. Fong JH, et al. Mol Biosyst. 2010 Oct;6(10):1821-8. doi: 10.1039/c005144f. Epub 2010 Jun 11. Mol Biosyst. 2010. PMID: 20544079 Free PMC article. - Dr. PIAS: an integrative system for assessing the druggability of protein-protein interactions.
Sugaya N, Furuya T. Sugaya N, et al. BMC Bioinformatics. 2011 Feb 9;12:50. doi: 10.1186/1471-2105-12-50. BMC Bioinformatics. 2011. PMID: 21303559 Free PMC article. - pKID Binds to KIX via an Unstructured Transition State with Nonnative Interactions.
Dahal L, Kwan TOC, Shammas SL, Clarke J. Dahal L, et al. Biophys J. 2017 Dec 19;113(12):2713-2722. doi: 10.1016/j.bpj.2017.10.016. Biophys J. 2017. PMID: 29262364 Free PMC article.
References
- Aloy P, Russell RB (2006) Structural systems biology: modelling protein interactions. Nat Rev Mol Cell Biol 7: 188–197 - PubMed
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases