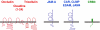Organization of multiprotein complexes at cell-cell junctions - PubMed (original) (raw)
Review
Organization of multiprotein complexes at cell-cell junctions
Klaus Ebnet. Histochem Cell Biol. 2008 Jul.
Abstract
The formation of stable cell-cell contacts is required for the generation of barrier-forming sheets of epithelial and endothelial cells. During various physiological processes like tissue development, wound healing or tumorigenesis, cellular junctions are reorganized to allow the release or the incorporation of individual cells. Cell-cell contact formation is regulated by multiprotein complexes which are localized at specific structures along the lateral cell junctions like the tight junctions and adherens junctions and which are targeted to these site through their association with cell adhesion molecules. Recent evidence indicates that several major protein complexes exist which have distinct functions during junction formation. However, this evidence also indicates that their composition is dynamic and subject to changes depending on the state of junction maturation. Thus, cell-cell contact formation and integrity is regulated by a complex network of protein complexes. Imbalancing this network by oncogenic proteins or pathogens results in barrier breakdown and eventually in cancer. Here, I will review the molecular organization of the major multiprotein complexes at junctions of epithelial cells and discuss their function in cell-cell contact formation and maintenance.
Figures
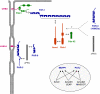
Fig. 1
Major protein complexes at adherens junctions. Two major protein complexes exist at AJs of epithelial cells. The cadherin–catenin complex consist of the Ca2+-dependent adhesion molecule E-cadherin and the armadillo repeat proteins p120ctn and β/γ-catenin which directly bind to the cytoplasmic domain of E-cadherin. α-catenin directly associates with β-catenin but not simultaneously with F-actin. The nectin–afadin complex consists of the Ca2+-independent adhesion molecule nectin and the PDZ protein afadin. Afadin contains a F-actin-binding domain and thus can link the nectin–afadin system to the actin cytoskeleton. The two adhesion complexes can be linked through several molecular interactions. Afadin can directly interact with α-catenin. It also interacts with ponsin/SH3P12 which can interact with the F-actin-binding protein vinculin, and it interacts with LMO7 and ADIP which both can interact with the F-actin binding protein α-actinin. The nature of the link between the cadherin–catenin complex and the actin cytoskeleton is still unclear. Double arrows indicate direct interactions, the question mark symbolizes the missing link
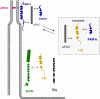
Fig. 2
Integral membrane proteins at tight junctions of epithelial and endothelial can be grouped into three classes based on their overall organizations. The first class is characterized by two extracellular loops, four transmembrane regions, and two cytoplasmic tails (occludin, claudins, tricellulin). The second class consists of Ig-SF members which all contain two Ig-like domains. The third class (contains only one member, CRB3) is characterized by a short extracellular domain (36 AA), a single transmembrane domain and a short cytoplasmic tail. In contrast to the other integral membrane proteins, the function of the extracellular domain of CRB3 is not clear
Fig. 3
Major protein complexes and functional classes of molecules at tight junctions. The TJs contain three major multi-protein complexes consisting largely of scaffolding proteins, the ZO protein complex, the CRB3–Pals1–PATJ complex and the PAR-3–aPKC–PAR-6 complex. Besides these three protein complexes which seem to be constitutively associated at TJs, a number of proteins with different functions has been identified at TJs. These include additional scaffolding proteins like MUPP1 and MAGI-1, adaptor proteins, transcription regulators and RNA processing factors, regulatory proteins like small GTPases and G-proteins, kinases and phosphatases, and heat shock proteins. Double arrows indicate direct interactions. Not all direct interactions that have been identified are depicted
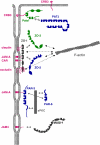
Fig. 4
Organization of the tight junctional plaque. The major protein complexes at TJs interact with specific transmembrane proteins. CRB3 recruits the Pals1–PATJ complex to TJs. CRB3 interacts with the PDZ domain of Pals1, Pals1 interacts with PATJ through a heterodimeric L27 domain interaction. CRB3 is also localized at the apical membrane domain of epithelial cells (Makarova et al. 2003). The CRB3–Pals1–PATJ complex regulates TJ formation but the mechanism is largely unknown. The ZO complex is associated with the membrane through multiple interactions of ZO-1 with various integral membrane proteins including occludin, claudins, JAM-A and CAR. ZO-2 and ZO-3 can interact with both ZO-1 and also with claudins. The ZO protein complex probably serves to link TJs to the cytoskeleton as all three ZO proteins directly interact with F-actin. The PAR-3–aPKC–PAR-6 complex is associated with the membrane through the interaction of PAR-3 with JAM-A. PAR-3 interacts with aPKC through its aPKC-interacting domain, PAR-6 interacts with aPKC through a PB1–PB1 domain interaction. A direct interaction between the PDZ domain of PAR-6 and PDZ domain 1 of PAR-3 has also been described. The PAR complex regulates the formation of TJs and apico-basal polarity. The JAM-A-related Ig-SF member JAM4 interacts directly with MAGI-1; the role of this protein complex is not clear. It should be noted that this drawing is incomplete as it does not depict interactions among the various protein complexes which have been described (e.g. PAR-6 can also associate with CRB3 and Pals1, ZO-3 can associate with PATJ). Also, the multiple PDZ domain protein MUPP1 (not depicted in this Figure, see Fig. 3 for a schematic representation) associates with claudins, JAM-A and CAR, as well as with angiomotin family members (Coyne et al. ; Hamazaki et al. ; Sugihara-Mizuno et al. 2007). Double arrows with solid lines indicate direct protein–protein interactions, double arrows with broken lines indicate interactions with F-actin
Fig. 5
A step-wise recruitment of proteins to cell–cell contacts. The earliest sites of stable physical interaction during cell–cell contact formation are primordial, spot-like AJs (pAJs) or puncta at the tips of cellular protrusions. During junctional maturation, the cellular protrusions of adjacent cells interdigitate, and multiple puncta are formed along the sides of protrusions. These puncta gradually fuse to form linear arrangements of cell–cell contacts sites thus generating a zipper-like appearance. During further maturation, cell–cell contacts are formed along the entire lateral cell surface, and the zipper-like cell–cell structure disappears. Cell contact-associated proteins are recruited in a step-wise manner. The pAJs/puncta are positive for integral membrane proteins (E-cadherin, JAM-A, nectin-2), but also peripheral membrane proteins (ZO-1, α-catenin, afadin) and contain proteins associated with AJs as well as TJs in polarized cells. During the formation of zipper-like cell contacts, occludin is recruited, probably through its interaction with ZO-1. Thereafter, PAR-3 is recruited by JAM-A and/or nectin-2, and claudins are recruited, probably through interaction with ZO-1. Indirect evidence suggests that aPKC and PAR-6 appear slightly later than PAR-3. The vertical bar reflects the increase in the contacting membrane area during cell–cell contact formation
Fig. 6
The Rich1–Amot complex at TJs. Rich1 is a RhoGAP with specificity for Cdc42 in epithelial cells. Rich1 interacts with Amot through a reciprocal BAR domain-dependent interaction, and Amot binding regulates Rich1 activity. Rich1 associates with Cdc42 through its GAP domain. Amot directly interacts with one (or several) of the PDZ domains 3–10 of PATJ that in turn associates with CRB3–Pals1. Surprisingly, Rich1 is directly or indirectly associated with PAR-3 and aPKC, and this PAR complex is distinct form the PAR-3–aPKC–PAR-6 complex because it does not contain PAR-6 and because aPKC cannot be associated with the 100 kDa isoform of PAR-3 which lacks the aPKC-binding domain. All three Amot-like proteins (Amot, Amotl1, Amotl2) form Rich1-independent protein complexes with MUPP1 and PATJ. Double arrows with solid lines indicate direct protein–protein interactions, arrows with broken lines indicate the presence of the two proteins in the same complex but the nature of the interaction has not been characterized in detail, yet
Fig. 7
The Scribble (Scrb), Discs large (Dlg) and Lethal giant larvae (Lgl) proteins localize to the basolateral membrane domain in polarized epithelial cells. Scrb and Lgl exist in a complex but it is not clear if the interaction is direct. Inset: During the process of cell–cell contact formation, Lgl forms a transient complex with aPKC and PAR-6 from which PAR-3 is excluded. After aPKC-induced dissociation of Lgl from the complex, PAR-3 associates with PAR-6 and aPKC which promotes TJ formation and the development of apical and basolateral membrane domains. Double arrows with solid lines indicate direct protein–protein interactions, double arrows with broken lines indicate the presence of the two proteins in the same complex but the nature of the interaction has not been characterized in detail, yet
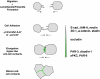
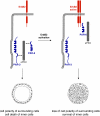
Fig. 8
The ErbB2 oncogene targets the PAR complex. In normal cells, PAR-3, aPKC and PAR-6 form a stable complex at TJs of epithelial cells (left panel). This complex is required for the fomation of TJs and the development of apico-basal polarity. ErbB2 activation triggers the association of the ErbB2 homodimer with PAR-6 and aPKC thereby disrupting the PAR-3–aPKC–PAR-6 complex. As a consequence, the development of apico-basal polarity is inhibited, and inner cells do not undergo apoptosis (right panel)
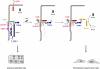
Fig. 9
TGFβ signaling targets the PAR complex at TJs. Left panel: Under normal conditions, TGFβ receptor I (TβRI) localizes to TJ through direct interactions with occludin and PAR-6. The cells maintain a polarized morrphology. Middle panel: TGFβ induces heterodimer formation of the two TGFβ receptors TβRI and TβR2 leading to activation of TβRII folllowed by TβRII-mediated phosphorylation of PAR-6 at Ser345. Right panel: Ser345-phosphorylated PAR-6 recruits Smurf1 leading to ubiquitination and degradation of the local pool of RhoA. As a consequence, the integrity of TJs is disturbed, the polarized morphology can not be maintained and the development of a fibroblastoid morphology is facilitated. In addition to PAR-6 phosphorylation, TGFβ signaling also induces downregulation of PAR-3. By targeting the PAR-3–aPKC–PAR-6 complex at TJs, TGFβ impairs the ability of cells to maintain a polarized morphology
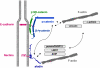
Fig. 10
Proteins at autotypic and heterotypic cell–cell contacts in the peripheral nervous system (PNS). A multitude of proteins localized at TJs of polarized epithelial cells localizes to autotypic glial–glial cell contacts and heteroytpic glial cell–axon contacts. Abbreviations: JXP juxtaparanodal region, MV microvilli, PNL paranodal loops, PNJ paranodal junction, SLI Schmidt-Lanterman incisure
Similar articles
- Molecular components of the adherens junction.
Niessen CM, Gottardi CJ. Niessen CM, et al. Biochim Biophys Acta. 2008 Mar;1778(3):562-71. doi: 10.1016/j.bbamem.2007.12.015. Epub 2008 Jan 14. Biochim Biophys Acta. 2008. PMID: 18206110 Free PMC article. Review. - The zonula adherens matura redefines the apical junction of intestinal epithelia.
Mangeol P, Massey-Harroche D, Sebbagh M, Richard F, Le Bivic A, Lenne PF. Mangeol P, et al. Proc Natl Acad Sci U S A. 2024 Feb 27;121(9):e2316722121. doi: 10.1073/pnas.2316722121. Epub 2024 Feb 20. Proc Natl Acad Sci U S A. 2024. PMID: 38377188 Free PMC article. - Nectin and afadin: novel organizers of intercellular junctions.
Takai Y, Nakanishi H. Takai Y, et al. J Cell Sci. 2003 Jan 1;116(Pt 1):17-27. doi: 10.1242/jcs.00167. J Cell Sci. 2003. PMID: 12456712 Review. - Cooperative role of nectin-nectin and nectin-afadin interactions in formation of nectin-based cell-cell adhesion.
Kurita S, Ogita H, Takai Y. Kurita S, et al. J Biol Chem. 2011 Oct 21;286(42):36297-303. doi: 10.1074/jbc.M111.261768. Epub 2011 Aug 31. J Biol Chem. 2011. PMID: 21880730 Free PMC article. - The blood-epididymis barrier and human male fertility.
Dubé E, Cyr DG. Dubé E, et al. Adv Exp Med Biol. 2012;763:218-36. doi: 10.1007/978-1-4614-4711-5_11. Adv Exp Med Biol. 2012. PMID: 23397627
Cited by
- Genetic ablation of afadin causes mislocalization and deformation of Paneth cells in the mouse small intestinal epithelium.
Tanaka-Okamoto M, Itoh Y, Miyoshi J, Mizoguchi A, Mizutani K, Takai Y, Inoue M. Tanaka-Okamoto M, et al. PLoS One. 2014 Oct 21;9(10):e110549. doi: 10.1371/journal.pone.0110549. eCollection 2014. PLoS One. 2014. PMID: 25333284 Free PMC article. - Stationed or Relocating: The Seesawing EMT/MET Determinants from Embryonic Development to Cancer Metastasis.
Li CH, Hsu TI, Chang YC, Chan MH, Lu PJ, Hsiao M. Li CH, et al. Biomedicines. 2021 Sep 18;9(9):1265. doi: 10.3390/biomedicines9091265. Biomedicines. 2021. PMID: 34572451 Free PMC article. Review. - Galangin and Kaempferol Alleviate the Indomethacin-Caused Cytotoxicity and Barrier Loss in Rat Intestinal Epithelial (IEC-6) Cells Via Mediating JNK/Src Activation.
Fan J, Zhao XH, Zhao JR, Li BR. Fan J, et al. ACS Omega. 2021 May 29;6(23):15046-15056. doi: 10.1021/acsomega.1c01167. eCollection 2021 Jun 15. ACS Omega. 2021. PMID: 34151085 Free PMC article. - A-kinase anchoring protein 2 is required for calcitonin-mediated invasion of cancer cells.
Thakkar A, Aljameeli A, Thomas S, Shah GV. Thakkar A, et al. Endocr Relat Cancer. 2016 Jan;23(1):1-14. doi: 10.1530/ERC-15-0425. Epub 2015 Oct 2. Endocr Relat Cancer. 2016. PMID: 26432469 Free PMC article. - Intestinal Barrier Function in Health and Disease-Any role of SARS-CoV-2?
Sharma L, Riva A. Sharma L, et al. Microorganisms. 2020 Nov 6;8(11):1744. doi: 10.3390/microorganisms8111744. Microorganisms. 2020. PMID: 33172188 Free PMC article. Review.
References
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PMC', 'value': 'PMC2133977', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC2133977/'}, {'type': 'PubMed', 'value': '8991100', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/8991100/'}\]}
- Adams CL, Nelson WJ, Smith SJ (1996) Quantitative analysis of cadherin-catenin-actin reorganization during development of cell–cell adhesion. J Cell Biol 135:1899–1911 - PMC - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PMC', 'value': 'PMC3369828', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC3369828/'}, {'type': 'PubMed', 'value': '12775840', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12775840/'}\]}
- Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S (2003) Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430–1434 - PMC - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '9299162', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9299162/'}\]}
- Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A (1997) Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res 235:374–384 - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '17060907', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17060907/'}\]}
- Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK (2006) Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 8:1235–1245 - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '12446711', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12446711/'}\]}
- Asada M, Irie K, Morimoto K, Yamada A, Ikeda W, Takeuchi M, Takai Y (2003) ADIP, a novel Afadin- and alpha-actinin-binding protein localized at cell–cell adherens junctions. J Biol Chem 278:4103–4111 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources