Gene rearrangement analysis and ancestral order inference from chloroplast genomes with inverted repeat - PubMed (original) (raw)
Comparative Study
Gene rearrangement analysis and ancestral order inference from chloroplast genomes with inverted repeat
Feng Yue et al. BMC Genomics. 2008.
Abstract
Background: Genome evolution is shaped not only by nucleotide substitutions, but also by structural changes including gene and genome duplications, insertions, deletions and gene order rearrangements. The most popular methods for reconstructing phylogeny from genome rearrangements include GRAPPA and MGR. However these methods are limited to cases where equal gene content or few deletions can be assumed. Since conserved duplicated regions are present in many chloroplast genomes, the inference of inverted repeats is needed in chloroplast phylogeny analysis and ancestral genome reconstruction.
Results: We extend GRAPPA and develop a new method GRAPPA-IR to handle chloroplast genomes. A test of GRAPPA-IR using divergent chloroplast genomes from land plants and green algae recovers the phylogeny congruent with prior studies, while analysis that do not consider IR structure fail to obtain the accepted topology. Our extensive simulation study also confirms that GRAPPA has better accuracy then the existing methods.
Conclusions: Tests on a biological and simulated dataset show GRAPPA-IR can accurately recover the genome phylogeny as well as ancestral gene orders. Close analysis of the ancestral genome structure suggests that genome rearrangement in chloroplasts is probably limited by inverted repeats with a conserved core region. In addition, the boundaries of inverted repeats are hot spots for gene duplications or deletions. The new GRAPPA-IR is available from http://phylo.cse.sc.edu.
Figures
Figure 1
Gene map of the Gossypium hirsutum chloroplast genome. The bright thick lines indicate the extent of the inverted repeats (IRa and IRb), which separate the genome into small (SSC) and large (LSC) single copy regions. The gene map is drawn using CGView [35].
Figure 2
The reference phylogeny of chloroplast genomes from land plants and green algae.
Figure 3
The best tree obtained by GRAPPA-IR. The topology is the same as the reference tree.
Figure 4
The best tree when no IR boundary is imposed, with score 73. Notice that the topology is different from the reference tree.
Figure 5
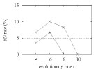
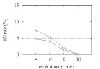
FN rate for GRAPPA-IR (solid line) and GRAPPA (dashed line) as a function of the evolutionary rate r for 6 genomes. The horizontal line indicates the 5% error level, a typical threshold of acceptability for accurate phylogenetic reconstruction [36].
Figure 6
FN rate for GRAPPA-IR (solid line) and GRAPPA (dashed line) as a function of the evolutionary rate r for 10 genomes.
Figure 7
Estimated gene contents for each region (only IR and SSC are shown).
Figure 8
Revised estimation of gene contents for each region (only IR and SSC are shown).
Figure 9
The scoring procedure of GRAPPA-IR when the tree is given.
Similar articles
- Phylogenetic reconstruction from transpositions.
Yue F, Zhang M, Tang J. Yue F, et al. BMC Genomics. 2008 Sep 16;9 Suppl 2(Suppl 2):S15. doi: 10.1186/1471-2164-9-S2-S15. BMC Genomics. 2008. PMID: 18831780 Free PMC article. - Molecular evolution of the plastid genome during diversification of the cotton genus.
Chen Z, Grover CE, Li P, Wang Y, Nie H, Zhao Y, Wang M, Liu F, Zhou Z, Wang X, Cai X, Wang K, Wendel JF, Hua J. Chen Z, et al. Mol Phylogenet Evol. 2017 Jul;112:268-276. doi: 10.1016/j.ympev.2017.04.014. Epub 2017 Apr 13. Mol Phylogenet Evol. 2017. PMID: 28414099 - Ancestral Genome Reconstruction on Whole Genome Level.
Feng B, Zhou L, Tang J. Feng B, et al. Curr Genomics. 2017 Aug;18(4):306-315. doi: 10.2174/1389202918666170307120943. Curr Genomics. 2017. PMID: 29081686 Free PMC article. Review.
Cited by
- Complete Chloroplast Genome of Medicinal Plant Lonicera japonica: Genome Rearrangement, Intron Gain and Loss, and Implications for Phylogenetic Studies.
He L, Qian J, Li X, Sun Z, Xu X, Chen S. He L, et al. Molecules. 2017 Feb 7;22(2):249. doi: 10.3390/molecules22020249. Molecules. 2017. PMID: 28178222 Free PMC article. - Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats.
Gao LZ, Liu YL, Zhang D, Li W, Gao J, Liu Y, Li K, Shi C, Zhao Y, Zhao YJ, Jiao JY, Mao SY, Gao CW, Eichler EE. Gao LZ, et al. Commun Biol. 2019 Jul 26;2:278. doi: 10.1038/s42003-019-0531-2. eCollection 2019. Commun Biol. 2019. PMID: 31372517 Free PMC article. - Phylogenetic relationships, selective pressure and molecular markers development of six species in subfamily Polygonoideae based on complete chloroplast genomes.
Feng Z, Zheng Y, Jiang Y, Pei J, Huang L. Feng Z, et al. Sci Rep. 2024 Apr 29;14(1):9783. doi: 10.1038/s41598-024-58934-7. Sci Rep. 2024. PMID: 38684694 Free PMC article. - Chloroplast genome sequence of Chongming lima bean (Phaseolus lunatus L.) and comparative analyses with other legume chloroplast genomes.
Tian S, Lu P, Zhang Z, Wu JQ, Zhang H, Shen H. Tian S, et al. BMC Genomics. 2021 Mar 18;22(1):194. doi: 10.1186/s12864-021-07467-8. BMC Genomics. 2021. PMID: 33736599 Free PMC article. - Insights into taxonomy and phylogenetic relationships of eleven Aristolochia species based on chloroplast genome.
Bai X, Wang G, Ren Y, Su Y, Han J. Bai X, et al. Front Plant Sci. 2023 Feb 13;14:1119041. doi: 10.3389/fpls.2023.1119041. eCollection 2023. Front Plant Sci. 2023. PMID: 36860895 Free PMC article.
References
- Muller K, Borsch T. Evolution of carnivory in Lentibulariaceae and the Lamiales. Plant Biol (Stuttg) 2004;6:477–490. - PubMed
- Gibbs R, Weinstock G, Metzker M, Muzny D, Sodergren E, Scherer S, Scott G, Steffen D, Worley K, Burch P. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:475–476. - PubMed
- Blanchette M, Kunisawa T, Sankoff D. Gene order breakpoint evidence in animal mitochondrial phylogeny. J Mol Evol. 1999;49:193–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources