Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis - PubMed (original) (raw)
Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis
Md Rowshon Alam et al. Nucleic Acids Res. 2008 May.
Abstract
We describe the synthesis and characterization of a 5' conjugate between a 2'-O-Me phosphorothioate antisense oligonucleotide and a bivalent RGD (arginine-glycine-aspartic acid) peptide that is a high-affinity ligand for the alphavbeta3 integrin. We used alphavbeta3-positive melanoma cells transfected with a reporter comprised of the firefly luciferase gene interrupted by an abnormally spliced intron. Intranuclear delivery of a specific antisense oligonucleotide (termed 623) corrects splicing and allows luciferase expression in these cells. The RGD-623 conjugate or a cationic lipid-623 complex produced significant increases in luciferase expression, while 'free' 623 did not. However, the kinetics of luciferase expression was distinct; the RGD-623 conjugate produced a gradual increase followed by a gradual decline, while the cationic lipid-623 complex caused a rapid increase followed by a monotonic decline. The subcellular distribution of the oligonucleotide delivered using cationic lipids included both cytoplasmic vesicles and the nucleus, while the RGD-623 conjugate was primarily found in cytoplasmic vesicles that partially co-localized with a marker for caveolae. Both the cellular uptake and the biological effect of the RGD-623 conjugate were blocked by excess RGD peptide. These observations suggest that the bivalent RGD peptide-oligonucleotide conjugate enters cells via a process of receptor-mediated endocytosis mediated by the alphavbeta3 integrin.
Figures
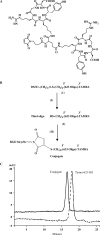
Figure 1.
(A) Chemical structure of the maleimide-bicyclic RGD peptide. The maleimide reactive group is positioned mid-way along a linker that joins the two cyclic RGD moieties. (B) Synthetic scheme for the conjugation of oligonucleotide 623 to the bivalent RGD peptide. The intermediates (1, 2) were purified after each step in the synthesis. (I) 100 mM DTT, 0.1 M TEAA buffer, 1% triethylamine; (II) maleimide-bicyclic-RGD peptide in H2O (1.5 equivalent), 400 mM KCl, 40% CH3CN, 3 h, RT. (C) HPLC analysis of the RGD peptide–oligonucleotide conjugate. HPLC analysis was performed as described in Materials and methods section. The elution profiles of the 5′-SH-3′-Tamra 623 oligonucleotide and its bivalent RGD conjugate are shown.
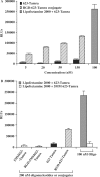
Figure 2.
Dose–response and specificity studies. (A) Cells were treated with either 623-Tamra, RGD–623-Tamra conjugate or 623-Tamra complexed with Lipofectamine 2000, as described in Materials and methods section, and luciferase activity was determined after 48 h and expressed as relative luminescence units (RLUs) per 1.5 × 105 cells. Black bars represent luciferase activity of 623-Tamra, patterned/striped bars represent the RGD–623-Tamra conjugate and the gray bar represents the activity of 100 nM 623-Tamra transfected using Lipofectamine 2000. (B) The effect of 623-Tamra or RGD–623-Tamra was compared to controls having five mismatched bases (indicated as 5MM623). The conjugates or free oligonucleotide were used at 200 nM while the Lipofectamine 2000 complexes were used at 100 nM oligonucleotide. Results (A and B) are the means and standard errors of three determinations.
Figure 3.
Time–response studies. Cells were treated with either 623-Tamra, RGD–623-Tamra conjugate or 623-Tamra complexed with Lipofectamine 2000, as described in Materials and methods section, and luciferase activity was determined at the times indicated. Black bars represent luciferase activity of 200 nM 623-Tamra, patterned/striped bars represent 200 nM RGD–623-Tamra conjugate and gray bars represent 100 nM 623-Tamra transfected using Lipofectamine 2000, all expressed as RLUs per 105 cells. Results are the means and standard errors of triplicate determinations.
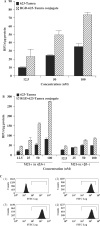
Figure 4.
(A) Total cell uptake. Cells in 12-well plates were treated with various concentrations of free 623-Tamra, or RGD–623-Tamra, for 4 h in OptiMEM at 37°C, then 1% FBS was added to each well. After 24 h of further treatment, the cells were rinsed three times in buffered saline solution and then lysed. Total cell-associated Tamra was measured using a Nanodrop® fluorimeter as described in Materials and methods section. Results represent means and standard errors of triplicate determinations and are expressed as relative fluorescence units (RFUs) per microgram cell protein. (B) Uptake of RGD–623-Tamra depends on αvβ3. Both αvβ3-positive M21+ and αvβ3-negative M21− human melanoma cells were exposed to several concentrations of RGD–623-Tamra or 623-Tamra. Total cell-associated Tamra was measured using a Nanodrop® fluorimeter as described in Materials and methods section. Results represent means and standard errors of triplicate determinations and are expressed as RFUs per microgram cell protein. (C) Lack of down-regulation of αvβ3 by RGD–623. A375SM-Luc705-B cells were maintained as controls or were treated with 200 nM RGD–623-Tamra or 623-Tamra for 24 h; thereafter, αvβ3 levels were determined by immunostaining with anti- αvβ3 and flow cytometry as described in Materials and methods section. (1) Cells not stained with primary anti- αvβ3; (2) control cells stained with anti- αvβ3, (3) cells treated with 200 nM RGD–623-Tamra and stained with anti- αvβ3, (4) cells treated with 200 nM 623-Tamra and stained with anti- αvβ3. Ordinate, number of cells; abcissa, log fluorescence intensity.
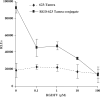
Figure 5.
Inhibition of effect with excess RGD peptide. Free RGDfV peptide at the indicated concentrations was added to the cells 30 min prior to treatment with either 623-Tamra or RGD-623-Tamra conjugate, as described in Materials and methods section. Luciferase activity was determined after 48 h. The dotted line represents luciferase activity of 200 nM 623-Tamra, and the solid line represents 200 nM RGD–623-Tamra conjugate. Results are the means and standard errors of triplicate determinations.
Figure 6.
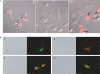
(A) Subcellular distribution of the conjugate. A375SM-Luc705-B cells were treated with either 623-Tamra, RGD–623-Tamra conjugate (both 200 nM) or with 623-Tamra complexed with Lipofectamine 2000 (100 nM), as described in Materials and methods section. Cells were observed using confocal fluorescence microscopy. Care was taken to make sure images were acquired from optical sections within the cell. Panel (1) RGD–623-Tamra conjugate, Panel (2) 623-Tamra, Panel (3) 623-Tamra complexed with Lipofectamine 2000. Images shown are overlaps of the Tamra-fluorescence and phase contrast images and are typical of multiple observations. In (1) white arrows indicate the position of the nucleus, while in (3) black arrows indicate nuclei that have accumulated Tamra-oligonucleotide. (B) Co-localization with endosomal pathway markers. RGD–623-Tamra oligonucleotide conjugate (100 nM) was co-incubated with: (1) Transferrin–Alexa 488 (200 nM) for 2 h: (2) Transferrin–Alexa 488 (200 nM) for 24 h; (3) Dextran–Alexa 488 (2 μM) for 2 h; (4) Dextran–Alexa 488 (2 μM) for 24 h. Live cells were observed by differential interference contrast (DIC) and confocal fluorescence microscopy as described in Materials and methods section. Concentrations of Alexa 488 transferrin and dextran were chosen to provide brightness similar to the Tamra conjugate. Panel (1) shows no overlap of RGD–623-Tamra (red) and Transferrin–Alexa 488 (green) at 2 h. Panel (3) shows substantial overlap (yellow-orange) of RGD–623-Tamra and Dextran–Alexa 488 at 2 h. Panels (2) and (4) show substantial overlap of RGD–623-Tamra with both Alexa 488 markers (yellow-orange areas) after 24 h.
Figure 7.
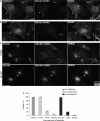
Co-localization with markers of endomembrane compartments and effects of inhibitors. Cells were treated with 50 nM RGD–623-Tamra for various periods and were then fixed, permeabilized and stained with antibodies to markers of various sub-cellular compartments, followed by Alexa 488 secondary antibody, as described in Materials and methods section. Fletches and boxes indicate areas of co-localization; boxed areas are expanded at higher magnification. (A) Co-localization with caveolin-1 at 6 h. (B) Co-localization with αvβ3 at 6 h. Note that similar patterns were observed during the period 2–6 h. (C) Co-localization with a trans-Golgi marker (TGN 230) at 6 or 24 h. (D) Cells were treated with cytochalasin D or β-cyclodextrin at the indicated concentrations for 15 min and then 100 nM RGD–623-Tamra was added. Total cell uptake after 4 h was measured using a Nanodrop® fluorimeter as described in Materials and methods section. No loss of cell viability was detected at the concentrations used, although the highest concentration of cytochalasin D caused some cell rounding. Results represent means and standard errors of triplicate determinations and are normalized based on cells receiving no inhibitor as 100%.
Figure 8.
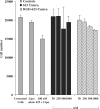
Short-term toxicity of 623–RGD conjugates. Cells were treated with either 623-Tamra, 623-Tamra complexed with Lipofectamine 2000 or RGD–623-Tamra conjugate, as described in Materials and methods section, and viable cell numbers were determined after 48 h by using a particle counter. Gray bars represent cell numbers of various controls i.e. cells treated with Lipofectamine 2000 (Lipo), and 623-Tamra complexed with Lipofectamine 2000. Black bars represent cell numbers for 623-Tamra, and patterned/striped bars represent RGD–623-Tamra conjugate, all at the indicated concentrations. Results are means and standard errors of three determinations.
Similar articles
- Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor.
Ming X, Alam MR, Fisher M, Yan Y, Chen X, Juliano RL. Ming X, et al. Nucleic Acids Res. 2010 Oct;38(19):6567-76. doi: 10.1093/nar/gkq534. Epub 2010 Jun 15. Nucleic Acids Res. 2010. PMID: 20551131 Free PMC article. - RGD Conjugated Dendritic Polylysine for Cellular Delivery of Antisense Oligonucleotide.
Le TD, Nakagawa O, Fisher M, Juliano RL, Yoo H. Le TD, et al. J Nanosci Nanotechnol. 2017 Apr;17(4):2353-357. doi: 10.1166/jnn.2017.13335. J Nanosci Nanotechnol. 2017. PMID: 29641161 - Targeted delivery of a splice-switching oligonucleotide by cationic polyplexes of RGD-oligonucleotide conjugate.
Ming X, Feng L. Ming X, et al. Mol Pharm. 2012 May 7;9(5):1502-10. doi: 10.1021/mp300113c. Epub 2012 Apr 25. Mol Pharm. 2012. PMID: 22497548 Free PMC article. - Radiolabeled multimeric cyclic RGD peptides as integrin alphavbeta3 targeted radiotracers for tumor imaging.
Liu S. Liu S. Mol Pharm. 2006 Sep-Oct;3(5):472-87. doi: 10.1021/mp060049x. Mol Pharm. 2006. PMID: 17009846 Review. - RGD peptide-based non-viral gene delivery vectors targeting integrin αvβ3 for cancer therapy.
Fu S, Xu X, Ma Y, Zhang S, Zhang S. Fu S, et al. J Drug Target. 2019 Jan;27(1):1-11. doi: 10.1080/1061186X.2018.1455841. Epub 2018 Apr 6. J Drug Target. 2019. PMID: 29564914 Review.
Cited by
- Viral Mimicry as a Design Template for Nucleic Acid Nanocarriers.
de la Fuente IF, Sawant SS, Tolentino MQ, Corrigan PM, Rouge JL. de la Fuente IF, et al. Front Chem. 2021 Mar 10;9:613209. doi: 10.3389/fchem.2021.613209. eCollection 2021. Front Chem. 2021. PMID: 33777893 Free PMC article. Review. - Oligonucleotide conjugates for therapeutic applications.
Winkler J. Winkler J. Ther Deliv. 2013 Jul;4(7):791-809. doi: 10.4155/tde.13.47. Ther Deliv. 2013. PMID: 23883124 Free PMC article. Review. - The delivery of therapeutic oligonucleotides.
Juliano RL. Juliano RL. Nucleic Acids Res. 2016 Aug 19;44(14):6518-48. doi: 10.1093/nar/gkw236. Epub 2016 Apr 15. Nucleic Acids Res. 2016. PMID: 27084936 Free PMC article. Review. - Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor.
Ming X, Alam MR, Fisher M, Yan Y, Chen X, Juliano RL. Ming X, et al. Nucleic Acids Res. 2010 Oct;38(19):6567-76. doi: 10.1093/nar/gkq534. Epub 2010 Jun 15. Nucleic Acids Res. 2010. PMID: 20551131 Free PMC article. - Unconventional internalization mechanisms underlying functional delivery of antisense oligonucleotides via cationic lipoplexes and polyplexes.
Ming X, Sato K, Juliano RL. Ming X, et al. J Control Release. 2011 Jul 15;153(1):83-92. doi: 10.1016/j.jconrel.2011.04.029. Epub 2011 May 4. J Control Release. 2011. PMID: 21571016 Free PMC article.
References
- Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. - PubMed
- Manoharan M. Oligonucleotide conjugates as potential antisense drugs with improved uptake, biodistribution, targeted delivery, and mechanism of action. Antisense Nucleic Acid Drug Dev. 2002;12:103–128. - PubMed
- Juliano RL, Yoo H. Aspects of the transport and delivery of antisense oligonucleotides. Curr. Opin. Mol. Ther. 2000;2:297–303. - PubMed
- Inoue A, Sawata SY, Taira K. Molecular design and delivery of siRNA. J. Drug. Target. 2006;14:448–455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 GM59299/GM/NIGMS NIH HHS/United States
- R21 CA121842/CA/NCI NIH HHS/United States
- U54CA119367/CA/NCI NIH HHS/United States
- P50 CA114747/CA/NCI NIH HHS/United States
- U54 CA119367/CA/NCI NIH HHS/United States
- P01 GM059299/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources