Lack of secondary structure characterizes the 5' ends of mammalian mitochondrial mRNAs - PubMed (original) (raw)
Lack of secondary structure characterizes the 5' ends of mammalian mitochondrial mRNAs
Christie N Jones et al. RNA. 2008 May.
Abstract
The mammalian mitochondrial genome encodes 13 proteins, which are synthesized at the direction of nine monocistronic and two dicistronic mRNAs. These mRNAs lack both 5' and 3' untranslated regions. The mechanism by which the specialized mitochondrial translational apparatus locates start codons and initiates translation of these leaderless mRNAs is currently unknown. To better understand this mechanism, the secondary structures near the start codons of all 13 open reading frames have been analyzed using RNA SHAPE chemistry. The extent of structure in these mRNAs as assessed experimentally is distinctly lower than would be predicted by current algorithms based on free energy minimization alone. We find that the 5' ends of all mitochondrial mRNAs are highly unstructured. The first 35 nucleotides for all mitochondrial mRNAs form structures with free energies less favorable than -3 kcal/mol, equal to or less than a single typical base pair. The start codons, which lie at the very 5' ends of these mRNAs, are accessible within single stranded motifs in all cases, making them potentially poised for ribosome binding. These data are consistent with a model in which the specialized mitochondrial ribosome preferentially allows passage of unstructured 5' sequences into the mRNA entrance site to participate in translation initiation.
Figures
FIGURE 1.
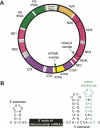
The mitochondrial genome and strategy for analyzing the 5′ ends of mitochondrial mRNAs. (A) The mammalian mitochondrial genome encodes 13 proteins, each of which is a subunit of the oxidative phosphorylation machinery. Protein coding genes and the two overlapping dicistronic mRNAs are shown explicitly. The protein encoding genes are generally separated by one or more tRNA genes (white); noncoding rRNA genes are green. (B) Messenger RNA fragments used for SHAPE analysis. The 5′ and 3′ extensions facilitate structural analysis of the 5′ end of the mRNA sequence and provide an efficient reverse transcriptase primer binding site, respectively.
FIGURE 2.
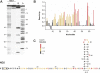
SHAPE analysis of the 5′ end of the ND2 mRNA. (A) Gel image showing the NMIA modification reaction (+), the no reagent control (−), and G and A dideoxy sequencing ladders. (B) Histogram of normalized SHAPE reactivities after correcting for drop off. The histogram bar colors correspond to SHAPE reactivities. Nucleotides are defined as having a high reactivity (red), moderate reactivity (yellow), or low reactivity (black). (C) Predicted secondary structure for the 5′ end of the ND2 mRNA as constrained by SHAPE reactivities. Nucleotide colors correspond to those in panel B. The reactivities for nucleotides 68–73 were assigned by visual analysis of the gel. Although the first 10 nt of this RNA have low (but still significant) reactivities, there are no obvious pairing partners for these positions in the RNA and flanking structure cassette. These nucleotides are thus predicted to be single stranded.
FIGURE 3.
SHAPE analysis of the 5′ end of the COIII mRNA. Panels A_–_C were generated as described for Figure 2 with the following additions. (A) Gel image showing sites of 2′-O-adduct formation for the COIII mRNA fragment both with and without the 5′ extension from the structure cassette. G and A sequencing ladders correspond to the COIII RNA containing the 5′ extension. (B) Absolute SHAPE reactivities. Histograms for the COIII mRNA fragment without and with the 5′ extension are blue and green, respectively. (C) Secondary structure model for the COIII mRNA fragment. Positions for which no data were obtained are gray. Identical structures are predicted for the RNAs without and with the 5′ extension.
FIGURE 4.
SHAPE analysis of the 5′ end of the COII mRNA. Panels A–C were generated as described for Figure 2 with the following additions. (A) Gel image shows sites of 2′-O-adduct formation for the 82-nt-long COII fragment and for the full-length mRNA. (B) Histograms for the COII fragment and full-length mRNA reactivities are in blue and green, respectively. (C) Secondary structure model for the COII mRNA fragment.
FIGURE 5.
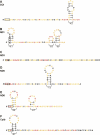
SHAPE-supported secondary structure models for the 5′ ends of the COI, ND1, ND3, ND5, ND6, and Cytb mRNAs. Nucleotides are colored by reactivity using the scheme shown in Figure 2. Calculated stabilities (Mathews et al. 2004) for individual stable stem–loop structures (in kcal/mol) are indicated. Start codons are boxed.
FIGURE 6.
Secondary structures at the 5′ translational start sites (A,C) and at the junctions between coding regions (B,D) for the dicistronic mRNAs. Start codons are boxed and the stop codons for the first cistrons (ND4L or ATP8) are underlined. Nucleotides are colored by SHAPE reactivity using the color scheme shown in Figure 2.
FIGURE 7.
Secondary structures for the leaderless mammalian mitochondrial mRNAs predicted without the use of experimental SHAPE information. Structures were predicted using RNAstructure (Mathews et al. 2004). Experimental SHAPE reactivities are superimposed in color using the scheme shown in Figure 2. Only those structures that differ from the SHAPE-constrained structures are shown.
Similar articles
- Mechanism of mRNA binding to bovine mitochondrial ribosomes.
Denslow ND, Michaels GS, Montoya J, Attardi G, O'Brien TW. Denslow ND, et al. J Biol Chem. 1989 May 15;264(14):8328-38. J Biol Chem. 1989. PMID: 2542274 - Preferential selection of the 5'-terminal start codon on leaderless mRNAs by mammalian mitochondrial ribosomes.
Christian BE, Spremulli LL. Christian BE, et al. J Biol Chem. 2010 Sep 3;285(36):28379-86. doi: 10.1074/jbc.M110.149054. Epub 2010 Jul 7. J Biol Chem. 2010. PMID: 20610392 Free PMC article. - Selection of initiator tRNA and start codon by mammalian mitochondrial initiation factor 3 in leaderless mRNA translation.
Lee M, Wakigawa T, Jia Q, Liu C, Huang R, Huang S, Nagao A, Suzuki T, Tomita K, Iwasaki S, Takeuchi-Tomita N. Lee M, et al. Nucleic Acids Res. 2025 Jan 24;53(3):gkaf021. doi: 10.1093/nar/gkaf021. Nucleic Acids Res. 2025. PMID: 39878211 Free PMC article. - Untranslated regions of mRNAs.
Mignone F, Gissi C, Liuni S, Pesole G. Mignone F, et al. Genome Biol. 2002;3(3):REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. Epub 2002 Feb 28. Genome Biol. 2002. PMID: 11897027 Free PMC article. Review. - Pushing the limits of the scanning mechanism for initiation of translation.
Kozak M. Kozak M. Gene. 2002 Oct 16;299(1-2):1-34. doi: 10.1016/s0378-1119(02)01056-9. Gene. 2002. PMID: 12459250 Free PMC article. Review.
Cited by
- Pentatricopeptide Protein PTCD2 Regulates COIII Translation in Mitochondria of the HeLa Cell Line.
Baleva MV, Chicherin I, Piunova U, Zgoda V, Patrushev MV, Levitskii S, Kamenski P. Baleva MV, et al. Int J Mol Sci. 2022 Nov 17;23(22):14241. doi: 10.3390/ijms232214241. Int J Mol Sci. 2022. PMID: 36430722 Free PMC article. - A small protein coded within the mitochondrial canonical gene nd4 regulates mitochondrial bioenergetics.
Kienzle L, Bettinazzi S, Choquette T, Brunet M, Khorami HH, Jacques JF, Moreau M, Roucou X, Landry CR, Angers A, Breton S. Kienzle L, et al. BMC Biol. 2023 May 18;21(1):111. doi: 10.1186/s12915-023-01609-y. BMC Biol. 2023. PMID: 37198654 Free PMC article. - Mitochondrial translation initiation machinery: conservation and diversification.
Kuzmenko A, Atkinson GC, Levitskii S, Zenkin N, Tenson T, Hauryliuk V, Kamenski P. Kuzmenko A, et al. Biochimie. 2014 May;100(100):132-40. doi: 10.1016/j.biochi.2013.07.024. Epub 2013 Aug 14. Biochimie. 2014. PMID: 23954798 Free PMC article. Review. - SHAPE-directed RNA secondary structure prediction.
Low JT, Weeks KM. Low JT, et al. Methods. 2010 Oct;52(2):150-8. doi: 10.1016/j.ymeth.2010.06.007. Epub 2010 Jun 8. Methods. 2010. PMID: 20554050 Free PMC article. Review. - Evaluating our ability to predict the structural disruption of RNA by SNPs.
Ritz J, Martin JS, Laederach A. Ritz J, et al. BMC Genomics. 2012 Jun 18;13 Suppl 4(Suppl 4):S6. doi: 10.1186/1471-2164-13-S4-S6. BMC Genomics. 2012. PMID: 22759654 Free PMC article.
References
- Anderson, S., Bankier, A.T., Barrell, B.G., de Bruijn, M.H.L., Coulson, A.R., Drouin, J., Eperon, I.C., Nierlich, D.P., Roe, B.A., Sanger, F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
- Anderson, S., de Brujin, M., Coulson, A., Eperon, I., Sanger, F., Young, I. Complete sequence of bovine mitochondrial DNA: Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982;156:683–717. - PubMed
- Badorrek, C.S., Weeks, K.M. Architecture of a γ retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–12672. - PubMed
- Boni, I.V. Diverse molecular mechanisms for translation initiation in prokaryotes. Mol. Biol. (Mosk.) 2006;40:658–668. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources