Reduced glioma growth following dexamethasone or anti-angiopoietin 2 treatment - PubMed (original) (raw)
Reduced glioma growth following dexamethasone or anti-angiopoietin 2 treatment
Jérôme Villeneuve et al. Brain Pathol. 2008 Jul.
Abstract
All patients with glioblastoma, the most aggressive and common form of brain cancer, develop cerebral edema. This complication is routinely treated with dexamethasone, a steroidal anti-inflammatory drug whose effects on brain tumors are not fully understood. Here we show that dexamethasone can reduce glioma growth in mice, even though it depletes infiltrating T cells with potential antitumor activity. More precisely, T cells with helper or cytotoxic function were sensitive to dexamethasone, but not those that were negative for the CD4 and CD8 molecules, including gammadelta and natural killer (NK) T cells. The antineoplastic effect of dexamethasone was indirect, as it did not meaningfully affect the growth and gene expression profile of glioma cells in vitro. In contrast, hundreds of dexamethasone-modulated genes, notably angiopoietin 2 (Angpt2), were identified in cultured cerebral endothelial cells by microarray analysis. The ability of dexamethasone to attenuate Angpt2 expression was confirmed in vitro and in vivo. Selective neutralization of Angpt2 using a peptide-Fc fusion protein reduced glioma growth and vascular enlargement to a greater extent than dexamethasone, without affecting T cell infiltration. In conclusion, this study suggests a mechanism by which dexamethasone can slow glioma growth, providing a new therapeutic target for malignant brain tumors.
Figures
Figure 1
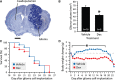
Dexamethasone reduces the growth of malignant gliomas in mice without increasing survival. A. Representative section of a glioma stained with thionin from a mouse killed 20 days after implantation of GL261 cells into the right caudoputamen. Scale bar, 1 mm. B. Stereological analysis revealed a 33% decrease in glioma volume in mice injected twice daily with 1 mg/kg dexamethasone (Dex) compared with sham‐treated mice. *Student's _t_‐test, P = 0.0008. Data show the mean ± SE. C. Kaplan–Meier curves showing no difference in the probability of survival between glioma‐bearing mice treated or not with dexamethasone (n = 13 per group). Log‐rank test, P = 0.68. D. A chronic reduction in body weight was recorded in glioma‐bearing mice treated with dexamethasone (n = 13 per group). *MANOVA with repeated measures (P < 0.0001) followed by Student's _t_‐tests (P < 0.02). The data show mean changes in body weight from the day of tumor implantation.
Figure 2
Dexamethasone depletes tumor‐infiltrating T cells, but not macrophages. A,B. T cells immunostained for CD3ε in glioma sections from mice injected twice daily for 20 days with vehicle or 1 mg/kg dexamethasone (Dex), respectively. Scale bar: 20 µm (A,B,D,E). C. Stereological analysis revealed a 51% decrease in the number of tumor‐infiltrating T cells in mice treated with dexamethasone. *Student's _t_‐test, P = 0.017. Data show the mean ± SE. D,E. Macrophages immunostained for Iba1 in glioma sections from mice treated with vehicle or dexamethasone, respectively. Note that macrophage morphology varies among regions (compare D with E), but that no difference was noted between the treatments. F. Stereological analysis showed no intergroup difference in the number of tumor‐associated macrophages. Student's _t_‐test, P = 0.5. Data show the mean ± SE. NSD, not statistically different.
Figure 3
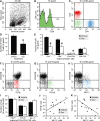
Dexamethasone selectively depletes tumor‐infiltrating T cells with helper or cytotoxic function. A–C. Flow cytometric analysis of cells isolated from a 20‐day glioma showing three predominant populations of CD3ε+ T cells. D. Cell counts revealed a 43% decrease in the overall number of CD3ε+ cells after treatment with dexamethasone (Dex; 1 mg/kg, twice daily). *Welch's _t_‐test, P = 0.0067. Data show the mean ± SE. E. All populations of CD3ε+ T cells were sensitive to dexamethasone, except CD4‐CD8‐ T cells. *Student's _t_‐test, P ≤ 0.02. Data show the mean ± SE. F–H. Phenotypic analysis showing CD3ε+CD4‐CD8‐ T cells positively labeled for T cell receptor (TCR) β (red), TCR γδ (green) and natural killer (NK)‐1.1 (blue). I. Cell counts showed no change in the number of double negative (DN) T cells expressing TCR β, TCR γδ or NK‐1.1 after dexamethasone treatment. Student's _t_‐test, P ≥ 0.14. Data show the mean ± SE. J,K. The number of double‐negative T cells labeled for NK‐1.1 correlated positively with that of TCR β+ cells (Pearson correlation, P = 0.022, R = 0.65) and negatively with that of TCR γδ+ cells (Pearson correlation, P = 0.016, R = −0.68).
Figure 4
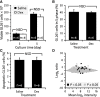
Cultured GL261 cells are virtually resistant to dexamethasone. A. As determined by flow cytometry, the number of viable cells (propidium iodide negative) did not differ between cultures supplemented or not with 1 µg/mL dexamethasone (Dex). Two‐way ANOVA (treatment, P = 0.23; time, P < 0.0001; interaction, P = 0.36). Data show the mean ± SE. NSD, not statistically different. B. No difference in the number of cells in the S phase of the cell cycle was detected after a 3‐day exposure to dexamethasone (Student's _t_‐test, P = 0.86). C. No difference in the number of apoptotic cells (annexin V positive and propidium iodide negative) was observed between the treatments (Student's _t_‐test, P = 0.31). D. Microarray analysis revealed minor changes in the transcriptional profile of GL261 cells after exposure to dexamethasone for 24 h. Each data point represents one of the 45 101 probe sets. Only 17 of these probe sets (black squares) had a _P_‐value <0.05 (see Table S3 for details). _x_‐axis, mean of the hybridization intensities of all samples; _y_‐axis, ratio of the mean hybridization intensities for dexamethasone‐treated cells versus control cells (n = 3 per group).
Figure 5
Dexamethasone slightly reduces tumor vascular density. A. Glioma section immunostained for CD31 showing morphological differences between the tumor and adjacent normal vasculature. The dashed line separates the tumor (top) from the nonneoplastic tissue (bottom). Scale bar: 200 µm. B,C. Higher magnification of capillaries in a glioma and the adjacent nonneoplastic tissue, respectively. Scale bar: 50 µm (B,C). D. Stereological analysis revealed a 14% decrease in tumor vascular density in mice treated twice daily with 1 mg/kg dexamethasone compared with mock‐treated mice. *Student's _t_‐test, P = 0.024. Data show the mean ± SE. E. No difference in tumor vessel caliber was recorded between mice treated or not with dexamethasone. Wilcoxon rank‐sum test, P = 0.2. Data show the mean ± SE. F,G. Glioma volume correlated better with vessel diameter (F, Pearson correlation, P = 0.041, R = 0.49) than with vascular volume (G, Pearson correlation, P = 0.053, R = 0.46). Each data point represents a mouse treated with dexamethasone or vehicle.
Figure 6
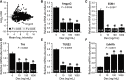
Genes modulated by dexamethasone in cultured cerebral endothelial cells. A. Microarray analysis revealed major changes in the transcriptional profile of bEnd.3 cells after exposure to dexamethasone for 24 h. Each data point represents one of the 45 101 probe sets; 1455 of these probe sets (black squares) had a _P_‐value <0.05 (see Table S4 for details). _x_‐axis, mean of the hybridization intensities of all samples; _y_‐axis, ratio of the mean hybridization intensities for dexamethasone‐treated cells versus control cells (n = 3 per group). B–F. Quantitative RT‐PCR analysis confirmed the differential expression of Angpt2, ESM1, Tnc, TGFβ2 and EdnRb mRNAs in bEnd.3 cells cultured for 24 h in the presence of different concentrations of dexamethasone (Dex) compared with mock‐treated cells. *Kruskal–Wallis test (P as indicated) followed by Dunn's test. The data (mean ± SE) are expressed as a ratio to 18S rRNA. Repeat analysis of Angpt2 with normalization to GAPDH mRNA instead of 18S rRNA gave similar results (data not shown).
Figure 7
Dexamethasone inhibits the endothelial expression of Angpt2 in gliomas, but not that of Tnc. A. Dark‐field photomicrograph showing in situ hybridization signals for Angpt2 mRNA in a 20‐day glioma. The dashed line separates the tumor (left) from the normal tissue (right). Scale bar: 500 µm. B. Bright‐field image at higher magnification of in situ hybridization signals for Angpt2 mRNA (black grains). Blue, thionin counterstaining. *Blood vessel. Scale bar: 20 µm (B,E). C. Double labeling for Angpt2 mRNA (in situ hybridization, black grains) and CD31 (immunohistochemistry, brown). *Blood vessel lumen. Scale bar: 20 µm (C,G). D. Stereological analysis revealed that the number of Angpt2 mRNA+ cells was approximately 40% lower in mice treated for 2 or 3 weeks with 1 mg/kg dexamethasone compared with sham‐treated mice. *Two‐way ANOVA (treatment, P = 0.0019; time, P = 0.065; interaction, P = 0.56) followed by Student's _t_‐test. Data show the mean ± SE. E. Optical density (OD) analysis of hybridization signals showed that the abundance of Angpt2 mRNA per positive cell was reduced by 38% at 2 weeks after dexamethasone treatment and by 27% at 3 weeks after dexamethasone treatment. *Two‐way ANOVA (treatment, P = 0.0016; time, P = 0.0058; interaction, P = 0.87) followed by Student's _t_‐test. Data show the mean ± SE. F. Bright‐field image of in situ hybridization signals for Tnc mRNA (black grains). Blue, thionin counterstaining. G. Double labeling for Tnc mRNA (in situ hybridization, black grains) and CD31 (immunohistochemistry, brown). Note that Tnc mRNA did not always colocalize with CD31 (arrows). *Blood vessel lumen. H. No intergroup difference in the number of Tnc mRNA+ cells was detected. Two‐way ANOVA (treatment, P = 0.42; time, P = 0.12; interaction, P = 0.99). Data show the mean ± SE. NSD, not statistically different.
Figure 8
Dexamethasone does not affect MMP2 and VEGFa expression in vivo, but reduces MMP2 expression in vitro. A,B. Quantitative RT‐PCR analysis showed reduced levels of MMP2 mRNA, but not of VEGFa mRNA, in bEnd.3 cells cultured for 24 h in the presence of different concentrations of dexamethasone (Dex) compared with mock‐treated cells. *Kruskal–Wallis test (P = 0.0173) followed by Dunn's test. Data (mean ± SE) are expressed as a ratio to 18S rRNA. C. Dark‐field photomicrograph of in situ hybridization signals for MMP2 mRNA in a 2‐week glioma. Note that MMP2 expression is not restricted to the endothelium, but is widespread in the tumor. Scale bar: 1 mm. D. Bright‐field photomicrograph of in situ hybridization signals for VEGFa mRNA (black grains) in a 3‐week glioma. Blue, thionin counterstaining. Scale bar: 20 µm. E. Optical densitometric analysis revealed no difference in MMP2 mRNA levels in mice treated or not with 1 mg/kg dexamethasone. *Two‐way ANOVA (treatment, P = 0.24; time, P < 0.0001; interaction, P = 0.79). Data show the mean ± SE. F. Stereological analysis revealed no difference in the number of VEGFa mRNA+ cells in mice treated or not with 1 mg/kg dexamethasone. *Two‐way ANOVA (treatment, P = 0.25; time, P = 0.06; interaction, P = 0.12). Data show the mean ± SE.
Figure 9
Specific neutralization of Angpt2 with L1‐10 reduces glioma growth and vascular enlargement without affecting T cell infiltration. A. Volumetric analysis showed a 50% decrease in glioma volume in L1‐10‐treated mice. *Wilcoxon rank‐sum test, P = 0.0002. Data show the mean ± SE. B. Increases in body weight were recorded at the beginning and at the end of experimentation in mice injected with L1‐10. *MANOVA with repeated measures (P = 0.002) followed by Student's _t_‐test (P < 0.05). The data show the mean change in body weight from the day of tumor implantation (n = 16 per group). C. Kaplan–Meier curves showing a modest increase in the probability of survival after treatment with dexamethasone. Log‐rank test, P = 0.005 (n = 13 per group). D. As determined by flow cytometry, the growth of GL261 cells in culture was not affected in the presence of 5 µM L1‐10. Two‐way ANOVA (treatment, P = 0.1; time, P < 0.0001; interaction, P = 0.07). Data show the mean ± SE. NSD, not statistically different. E. Stereological analysis revealed a 9% decrease in tumor vascular density in L1‐10‐treated mice. *Wilcoxon rank‐sum test, P = 0.03. Data show the mean ± SE. F,G. Tumor vessel diameter was reduced by 29% in L1‐10‐treated mice and correlated positively with glioma volume (Pearson correlation, P = 0.0002, R = 0.61). *Wilcoxon rank‐sum test, P = 0.0053. Each data point represents a mouse treated with L1‐10 or vehicle. H. No intergroup difference was detected in CD3+ T cell density. Wilcoxon rank‐sum test, P = 0.07. Data show the mean ± SE. I. A 17% decrease in the number of tumor‐associated Iba1+ macrophages was observed after L1‐10 treatment. Wilcoxon rank‐sum test, P = 0.004. Data show the mean ± SE.
Similar articles
- Dexamethasone alleviates tumor-associated brain damage and angiogenesis.
Fan Z, Sehm T, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan NE. Fan Z, et al. PLoS One. 2014 Apr 8;9(4):e93264. doi: 10.1371/journal.pone.0093264. eCollection 2014. PLoS One. 2014. PMID: 24714627 Free PMC article. - Dexamethasone-induced abolition of the inflammatory response in an experimental glioma model: a flow cytometry study.
Badie B, Schartner JM, Paul J, Bartley BA, Vorpahl J, Preston JK. Badie B, et al. J Neurosurg. 2000 Oct;93(4):634-9. doi: 10.3171/jns.2000.93.4.0634. J Neurosurg. 2000. PMID: 11014542 - Expression of angiopoietin-2 in human glioma cells and its role for angiogenesis.
Koga K, Todaka T, Morioka M, Hamada J, Kai Y, Yano S, Okamura A, Takakura N, Suda T, Ushio Y. Koga K, et al. Cancer Res. 2001 Aug 15;61(16):6248-54. Cancer Res. 2001. PMID: 11507079 - Novel anti-angiogenic therapies for malignant gliomas.
Norden AD, Drappatz J, Wen PY. Norden AD, et al. Lancet Neurol. 2008 Dec;7(12):1152-60. doi: 10.1016/S1474-4422(08)70260-6. Lancet Neurol. 2008. PMID: 19007739 Review. - Anti-VEGF therapies for malignant glioma: treatment effects and escape mechanisms.
Miletic H, Niclou SP, Johansson M, Bjerkvig R. Miletic H, et al. Expert Opin Ther Targets. 2009 Apr;13(4):455-68. doi: 10.1517/14728220902806444. Expert Opin Ther Targets. 2009. PMID: 19335067 Review.
Cited by
- Dexamethasone in Patients with Glioblastoma: A Systematic Review and Meta-Analysis.
Scheffler P, Fung C, Momjian S, Koessinger D, Häni L, Neidert N, Straehle J, Volz F, Schnell O, Beck J, El Rahal A. Scheffler P, et al. Cancers (Basel). 2024 Apr 1;16(7):1393. doi: 10.3390/cancers16071393. Cancers (Basel). 2024. PMID: 38611071 Free PMC article. Review. - Angiopoietin-2 interferes with anti-VEGFR2-induced vessel normalization and survival benefit in mice bearing gliomas.
Chae SS, Kamoun WS, Farrar CT, Kirkpatrick ND, Niemeyer E, de Graaf AM, Sorensen AG, Munn LL, Jain RK, Fukumura D. Chae SS, et al. Clin Cancer Res. 2010 Jul 15;16(14):3618-27. doi: 10.1158/1078-0432.CCR-09-3073. Epub 2010 May 25. Clin Cancer Res. 2010. PMID: 20501615 Free PMC article. - The evaluation of six genes combined value in glioma diagnosis and prognosis.
Lin P, He L, Tian N, Qi X. Lin P, et al. J Cancer Res Clin Oncol. 2023 Oct;149(13):12413-12433. doi: 10.1007/s00432-023-05082-6. Epub 2023 Jul 13. J Cancer Res Clin Oncol. 2023. PMID: 37439825 - Identification of a Dexamethasone Mediated Radioprotection Mechanism Reveals New Therapeutic Vulnerabilities in Glioblastoma.
Aldaz P, Auzmendi-Iriarte J, Durántez M, Lasheras-Otero I, Carrasco-Garcia E, Zelaya MV, Bragado L, Olías-Arjona A, Egaña L, Samprón N, Morilla I, Redondo-Muñoz M, Rico M, Squatrito M, Maria-Alonso M, Fernández-Irigoyen J, Santamaria E, Larráyoz IM, Wellbrock C, Matheu A, Arozarena I. Aldaz P, et al. Cancers (Basel). 2021 Jan 19;13(2):361. doi: 10.3390/cancers13020361. Cancers (Basel). 2021. PMID: 33478100 Free PMC article. - Crawling phagocytes recruited in the brain vasculature after pertussis toxin exposure through IL6, ICAM1 and ITGαM.
Richard JF, Roy M, Audoy-Rémus J, Tremblay P, Vallières L. Richard JF, et al. Brain Pathol. 2011 Nov;21(6):661-71. doi: 10.1111/j.1750-3639.2011.00490.x. Epub 2011 May 23. Brain Pathol. 2011. PMID: 21418369 Free PMC article.
References
- Ashwell JD, Lu FW, Vacchio MS (2000) Glucocorticoids in t cell development and function. Annu Rev Immunol 18:309–345. - PubMed
- Benedetti S, Pirola B, Poliani PL, Cajola L, Pollo B, Bagnati R et al (2003) Dexamethasone inhibits the anti‐tumor effect of interleukin 4 on rat experimental gliomas. Gene Ther 10:188–192. - PubMed
- Brooks WH, Markesbery WR, Gupta GD, Roszman TL (1978) Relationship of lymphocyte invasion and survival of brain tumor patients. Ann Neurol 4:219–224. - PubMed
- Carding SR, Egan PJ (2002) Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol 2:336–345. - PubMed
- Chiquet‐Ehrismann R, Chiquet M (2003) Tenascins: regulation and putative functions during pathological stress. J Pathol 200:488–499. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials