Gene flow persists millions of years after speciation in Heliconius butterflies - PubMed (original) (raw)
Gene flow persists millions of years after speciation in Heliconius butterflies
Marcus R Kronforst. BMC Evol Biol. 2008.
Abstract
Background: Hybridization, or the interbreeding of two species, is now recognized as an important process in the evolution of many organisms. However, the extent to which hybridization results in the transfer of genetic material across the species boundary (introgression) remains unknown in many systems, as does the length of time after initial divergence that the species boundary remains porous to such gene flow.
Results: Here I use genome-wide genotypic and DNA sequence data to show that there is introgression and admixture between the melpomene/cydno and silvaniform clades of the butterfly genus Heliconius, groups that separated from one another as many as 30 million generations ago. Estimates of historical migration based on 523 DNA sequences from 14 genes suggest unidirectional gene flow from the melpomene/cydno clade into the silvaniform clade. Furthermore, genetic clustering based on 520 amplified fragment length polymorphisms (AFLPs) identified multiple individuals of mixed ancestry showing that introgression is on-going.
Conclusion: These results demonstrate that genomes can remain porous to gene flow very long after initial divergence. This, in turn, greatly expands the evolutionary potential afforded by introgression. Phenotypic and species diversity in a wide variety of organisms, including Heliconius, have likely arisen from introgressive hybridization. Evidence for continuous gene flow over millions of years points to introgression as a potentially important source of genetic variation to fuel the evolution of novel forms.
Figures
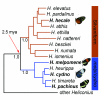
Figure 1
Bayesian phylogeny showing the relationship between the distantly-related silvaniform and melpomene/cydno clades of the genus Heliconius. Posterior probabilities for three major nodes are shown (1.0), as are the wing pattern phenotypes for the four analyzed species. The age of the split between the melpomene/cydno and silvaniform clades was estimated assuming an evolutionary rate of 1.15% per lineage per million years [32].
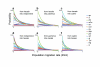
Figure 2
Posterior probability distributions for between-species population migration rates. Three pairwise comparisons based on DNA sequence data for 14 genes revealed non-zero rates of historical gene flow from the melpomene/cydno clade into the silvaniform clade (a, b, c). Locus specific estimates of gene flow into H. hecale from H. melpomene, H. pachinus, and H. cydno, respectively, for the two loci that exhibit evidence of between-clade introgression, cubitus interruptus and white (d, e, f).
Figure 3
Marginal posterior probability distributions for locus-specific migration rates. Probability distributions for gene flow from H. hecale into H. melpomene, H. pachinus, and H. cydno, respectively (a, b, c). Probability distributions for gene flow into H. hecale from H. melpomene, H. pachinus, and H. cydno, respectively (d, e, f). Despite substantial variation in distribution shapes, most peak at or near zero suggestive of little or no gene flow.
Figure 4
Gene genealogies for cubitus interruptus and white. Across loci, haplotypes from H. hecale and the melpomene/cydno clade were reciprocally monophyletic except for at cubitus interruptus and white [24]. The lack of association between species and tree topology for these two loci is indicative of interspecific gene flow. Nodes with posterior probabilities > 0.60 are labeled.
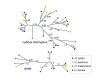
Figure 5
Genetic clustering based on 520 AFLP loci. Each individual is represented by a narrow vertical column with the proportion of the four colors indicating the genome proportion derived from each of the four populations. Two H. hecale and two H. cydno individuals exhibited evidence of mixed ancestry with the opposite clade (asterisks).
Similar articles
- Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies.
Kronforst MR, Young LG, Blume LM, Gilbert LE. Kronforst MR, et al. Evolution. 2006 Jun;60(6):1254-68. Evolution. 2006. PMID: 16892975 - Genome-wide patterns of divergence and gene flow across a butterfly radiation.
Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra KK, Davey JW, Baxter SW, Blaxter ML, Mallet J, Jiggins CD. Nadeau NJ, et al. Mol Ecol. 2013 Feb;22(3):814-26. doi: 10.1111/j.1365-294X.2012.05730.x. Epub 2012 Aug 25. Mol Ecol. 2013. PMID: 22924870 - Adaptive introgression across species boundaries in Heliconius butterflies.
Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD. Pardo-Diaz C, et al. PLoS Genet. 2012;8(6):e1002752. doi: 10.1371/journal.pgen.1002752. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737081 Free PMC article. - Review. Hybrid trait speciation and Heliconius butterflies.
Jiggins CD, Salazar C, Linares M, Mavarez J. Jiggins CD, et al. Philos Trans R Soc Lond B Biol Sci. 2008 Sep 27;363(1506):3047-54. doi: 10.1098/rstb.2008.0065. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 18579480 Free PMC article. Review. - Introgression of wing pattern alleles and speciation via homoploid hybridization in Heliconius butterflies: a review of evidence from the genome.
Brower AV. Brower AV. Proc Biol Sci. 2012 Dec 12;280(1752):20122302. doi: 10.1098/rspb.2012.2302. Print 2013 Feb 7. Proc Biol Sci. 2012. PMID: 23235702 Free PMC article. Review.
Cited by
- Introgression and rapid species turnover in sympatric damselflies.
Sánchez-Guillén RA, Wellenreuther M, Cordero-Rivera A, Hansson B. Sánchez-Guillén RA, et al. BMC Evol Biol. 2011 Jul 18;11:210. doi: 10.1186/1471-2148-11-210. BMC Evol Biol. 2011. PMID: 21767355 Free PMC article. - Genome-wide introgression among distantly related Heliconius butterfly species.
Zhang W, Dasmahapatra KK, Mallet J, Moreira GR, Kronforst MR. Zhang W, et al. Genome Biol. 2016 Feb 27;17:25. doi: 10.1186/s13059-016-0889-0. Genome Biol. 2016. PMID: 26921238 Free PMC article. - Butterfly genome reveals promiscuous exchange of mimicry adaptations among species.
Heliconius Genome Consortium. Heliconius Genome Consortium. Nature. 2012 Jul 5;487(7405):94-8. doi: 10.1038/nature11041. Nature. 2012. PMID: 22722851 Free PMC article. - A neutral view of the evolving genomic architecture of speciation.
Southcott L, Kronforst MR. Southcott L, et al. Ecol Evol. 2017 Jul 6;7(16):6358-6366. doi: 10.1002/ece3.3190. eCollection 2017 Aug. Ecol Evol. 2017. PMID: 28861239 Free PMC article. - Molecular evidence for hybridization in Colias (Lepidoptera: Pieridae): are Colias hybrids really hybrids?
Dwyer HE, Jasieniuk M, Okada M, Shapiro AM. Dwyer HE, et al. Ecol Evol. 2015 Jul;5(14):2865-77. doi: 10.1002/ece3.1574. Epub 2015 Jun 25. Ecol Evol. 2015. PMID: 26306172 Free PMC article.
References
- Rieseberg L. Hybrid origins of plant species. Annu Rev Ecol Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. - DOI
- Grant V. Plant Speciation. New York, Columbia Univ. Press; 1981.
- Mayr E. Animal Species and Evolution. Cambridge, Harvard Univ. Press; 1963.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources