Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation - PubMed (original) (raw)
Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation
Shaun D Fouse et al. Cell Stem Cell. 2008.
Abstract
We report here genome-wide mapping of DNA methylation patterns at proximal promoter regions in mouse embryonic stem (mES) cells. Most methylated genes are differentiation associated and repressed in mES cells. By contrast, the unmethylated gene set includes many housekeeping and pluripotency genes. By crossreferencing methylation patterns to genome-wide mapping of histone H3 lysine (K) 4/27 trimethylation and binding of Oct4, Nanog, and Polycomb proteins on gene promoters, we found that promoter DNA methylation is the only marker of this group present on approximately 30% of genes, many of which are silenced in mES cells. In demethylated mutant mES cells, we saw upregulation of a subset of X-linked genes and developmental genes that are methylated in wild-type mES cells, but lack either H3K4 and H3K27 trimethylation or association with Polycomb, Oct4, or Nanog. Our data suggest that in mES cells promoter methylation represents a unique epigenetic program that complements other regulatory mechanisms to ensure appropriate gene expression.
Figures
Figure 1. Using mDIP-CHIP Assay to Profile the Promoter Methylation Pattern in Wild Type mESCs
(A) All probes for the array are classified as either inside or within 50bp of a CpG island (white) or outside a CpG island (grey) and then grouped by their average log2 ratio. The solid line is placed at log2 ratio of +0.2 value to annotate the proportion of methylated gene promoters. (B) Using the average Log2 ratio, we selected 3 genes thought to be methylated as well as 3 that were thought to be unmethylated. For the bisulfite confirmation, each line represents an individual clone and each circle represents a CpG dinucleotide. The red line indicates the region we analyzed. Filled circles are methylated CpGs and open circles are unmethylated. (C) The average signals (Log2 ratio) for each probe (block with double arrowheads) in the promoter region are plotted in the UCSC genome browser – here we take the Trpc1 gene as an example. Note the dark probes annotate probes with high Log2 ratios whereas light color probes represent probes with low or negative Log2 ratios. (D) The confirmation of methylation status for the probes in the Trpc1 gene either inside or outside of a CpG island. Red bars (I-IV) indicate regions analyzed. Region I shows a gel picture of a McrBC-HhaI genomic PCR, indicating the unmethylated state of the CpG island of the Trpc1 transcription start site. II-IV show bisulfite genomic sequencing results of methylated probes upstream of the CpG islands, indicating a gradient of methylation towards the probes with high log2 ratios (filled dots are methylated CpG sites).
Figure 2. CpG Density of Methylated and Unmethylated Promoters
(A) Classification of all promoters, methylated promoters or unmethylated promoters with High (HCP), Intermediate (ICP) and Low (LCP) CpG content. It should be noted that these classifications were assigned for V6.5 mouse ES cells which are similar but no the same as the J1 mouse ES cells used in our study. (B) Breakdown of methylation status for HCP, ICP and LCP promoters. (C) Percentage of genes with methylated CpG islands in HCP and ICP containing genes. By definition, there are no CpG islands in LCP containing genes.
Figure 3. Comparision of Methylated and Unmethylated Genes with H3K4me3 and H3K27me3
Gene ontology classifications for Methylated (A) or Unmethylated (B) genes. The GO term is on the y-axis and the p value indicating significance of enrichment is on the x-axis.
Figure 4. Comparison of Methylated and Unmethylated Genes With H3K4me3 and H3K27me3 as Well as Nanog, Oct4, and PcG Bound Genes
(A) Classification of all promoters, methylated promoters or unmethylated promoters with H3K4me3, H3k27me3, H3K4me3 & H3K27me3 (bivalent) or neither mark. Methylated genes are enriched for genes without either histone mark. (B) Percentage of K4, K4/K27, K27 or neither bound genes that are HCP, ICP or LCP. (C) Breakdown of methylation status for genes with H3K4me3 only, H3k27me3 only, H3K4me3 & H3K27me3 (bivalent) or neither mark. (D) Bar graph showing the breakdown of methylation status for PcG, Nanog or Oct4 bound genes in mESCs. The Nanog, Oct4 and PcG bound genes that are either methylated or unmethyalted are listed in Table S1.
Figure 5. Gene Expression Profiling of DNA Methylation Deficient mESCs
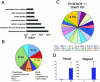
(A) Gene ontology analysis for up-regulated genes in TKO cells shows enrichment of transcription factor activity, protein binding, extracellular region, and developmental genes. (B) Tissue specificity of over expression genes in TKO cells. Up-regulated genes were analyzed for the tissues they are normally expressed in using the GNF database. Note the over-representation of genes that are expressed in reproductive tissues including ovary/testis (16.2%) and placenta/umbilical cord (11.3%). (C) The chromosomal location of the 390 genes that are upregulated in TKO mESC compared to wild type mESC (clockwise from Chromosome 1 to X and Y sex chromosomes). The percentage of genes upregulated on the X chromosome is 14.5%. (D) Confirmation of Rhox2 and Magea3 that are upregulated in DNA methylation deficient mESCs by Q-PCR analysis. * P <0.05
Figure 6. Comparison of Upregulated Genes in Demethylated mESCs With Oct4, Nanog and PcG Bound Genes
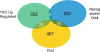
Venn diagram showing the minimal overlap between the TKO upregulated genes (green), Nanog and/or Oct4 bound genes (within 10kb proximal promoter region) (blue), and PcG bound genes (yellow). It should be noted that there is one gene, Podxl, which overlaps with all three categories. This analysis includes all genes, not just those present on the promoter methylation array. The Nanog, Oct4 (Loh et al., 2006) and PcG (Boyer et al., 2006) genes that overlap with TKO upregulated genes are listed in Table S7.
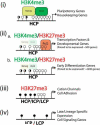
Figure 7. A Schematic Summary of Epigenetic and Transcriptional Regulation in mESCs
The patterns of DNA methylation and histone modifications are depicted into four subclasses of gene promoters with H3K4me3 (i), bivalent genes (ii) with (a.) or without (b.) polycomb binding, H3K27me3 only (iii), and no histone marks (iv). Gene promoters are further annotated as HCP, ICP, and LCP that corresponds to different density of CpG dinucleotides and DNA methylation. Open circles designate unmethylated CpG sites, filled circles represent methylated CpG sites and half filled circles represent promoters which can be found either methylated or unmethylated. Unmethylated CpG islands are enriched in HCP promoters with H3K4me only or bivalent H3K4/K27 marks. Methylated CpGs are enriched in LCP and ICP gene promoters without H3 K4 and K27 trimethylation or with K27me3 only. The preferential interaction of Polycomb proteins and Oct4/Nanog complex with different classes of gene promoter is also illustrated. Different classes of genes that are either expressed or repressed in mESCs are listed next to each type of promoters.
Similar articles
- Dynamic epigenetic regulation of the Oct4 and Nanog regulatory regions during neural differentiation in rhesus nuclear transfer embryonic stem cells.
Wang K, Chen Y, Chang EA, Knott JG, Cibelli JB. Wang K, et al. Cloning Stem Cells. 2009 Dec;11(4):483-96. doi: 10.1089/clo.2009.0019. Cloning Stem Cells. 2009. PMID: 20025521 - The polycomb group protein Suz12 is required for embryonic stem cell differentiation.
Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. Pasini D, et al. Mol Cell Biol. 2007 May;27(10):3769-79. doi: 10.1128/MCB.01432-06. Epub 2007 Mar 5. Mol Cell Biol. 2007. PMID: 17339329 Free PMC article. - Polycomb protein EED is required for silencing of pluripotency genes upon ESC differentiation.
Obier N, Lin Q, Cauchy P, Hornich V, Zenke M, Becker M, Müller AM. Obier N, et al. Stem Cell Rev Rep. 2015 Feb;11(1):50-61. doi: 10.1007/s12015-014-9550-z. Stem Cell Rev Rep. 2015. PMID: 25134795 - Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs.
Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Kashyap V, et al. Stem Cells Dev. 2009 Sep;18(7):1093-108. doi: 10.1089/scd.2009.0113. Stem Cells Dev. 2009. PMID: 19480567 Free PMC article. Review. - Pluripotency and Epigenetic Factors in Mouse Embryonic Stem Cell Fate Regulation.
Morey L, Santanach A, Di Croce L. Morey L, et al. Mol Cell Biol. 2015 Aug;35(16):2716-28. doi: 10.1128/MCB.00266-15. Epub 2015 Jun 1. Mol Cell Biol. 2015. PMID: 26031336 Free PMC article. Review.
Cited by
- Chromatin modifications and genomic contexts linked to dynamic DNA methylation patterns across human cell types.
Yan H, Zhang D, Liu H, Wei Y, Lv J, Wang F, Zhang C, Wu Q, Su J, Zhang Y. Yan H, et al. Sci Rep. 2015 Feb 12;5:8410. doi: 10.1038/srep08410. Sci Rep. 2015. PMID: 25673498 Free PMC article. - Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation.
Wu H, Zhang Y. Wu H, et al. Genes Dev. 2011 Dec 1;25(23):2436-52. doi: 10.1101/gad.179184.111. Genes Dev. 2011. PMID: 22156206 Free PMC article. Review. - Transcriptional competence in pluripotency.
Ram EV, Meshorer E. Ram EV, et al. Genes Dev. 2009 Dec 15;23(24):2793-8. doi: 10.1101/gad.1881609. Genes Dev. 2009. PMID: 20008929 Free PMC article. - Epigenetic control of embryonic stem cell fate.
Christophersen NS, Helin K. Christophersen NS, et al. J Exp Med. 2010 Oct 25;207(11):2287-95. doi: 10.1084/jem.20101438. J Exp Med. 2010. PMID: 20975044 Free PMC article. Review. - Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells.
Klein D, Benchellal M, Kleff V, Jakob HG, Ergün S. Klein D, et al. Sci Rep. 2013 Oct 22;3:2178. doi: 10.1038/srep02178. Sci Rep. 2013. PMID: 24145756 Free PMC article.
References
- Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van Oostwaard D, Andrews PW, Stojkovic M, Smith N, et al. Restriction Landmark Genome Scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. - PubMed
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1F31MH070204/MH/NIMH NIH HHS/United States
- NS051411/NS/NINDS NIH HHS/United States
- F31 MH070204/MH/NIMH NIH HHS/United States
- NS044405/NS/NINDS NIH HHS/United States
- R01 NS051411/NS/NINDS NIH HHS/United States
- R56 NS051411/NS/NINDS NIH HHS/United States
- R01 NS044405/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous