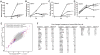MicroRNA regulation of cell lineages in mouse and human embryonic stem cells - PubMed (original) (raw)
MicroRNA regulation of cell lineages in mouse and human embryonic stem cells
Kathryn N Ivey et al. Cell Stem Cell. 2008.
Abstract
Cell fate decisions of pluripotent embryonic stem (ES) cells are dictated by activation and repression of lineage-specific genes. Numerous signaling and transcriptional networks progressively narrow and specify the potential of ES cells. Whether specific microRNAs help refine and limit gene expression and, thereby, could be used to manipulate ES cell differentiation has largely been unexplored. Here, we show that two serum response factor (SRF)-dependent muscle-specific microRNAs, miR-1 and miR-133, promote mesoderm formation from ES cells but have opposing functions during further differentiation into cardiac muscle progenitors. Furthermore, miR-1 and miR-133 were potent repressors of nonmuscle gene expression and cell fate during mouse and human ES cell differentiation. miR-1's effects were in part mediated by translational repression of the Notch ligand Delta-like 1 (Dll-1). Our findings indicate that muscle-specific miRNAs reinforce the silencing of nonmuscle genes during cell lineage commitment and suggest that miRNAs may have general utility in regulating cell-fate decisions from pluripotent ES cells.
Figures
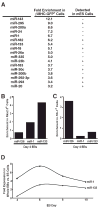
Figure 1. Identification of miRNAs expressed in ES cell–derived cardiomyocytes
(A) mES cells carrying a GFP transgene under control of the cardiomyocyte-specific β-myosin heavy chain promoter were differentiated for 13 days as embryoid bodies (EBs), sorted by GFP expression, and analyzed by miRNA microarray. miRNAs enriched at least threefold in the GFP+ compared to GFP− cell populations are listed along with their fold enrichment and whether they were detected in ES cells. (B, C) Quantitative RT-PCR (qRT-PCR) showing enrichment of miR-1 and miR-133 inflow-sorted Nkx2.5-GFP+ cardiac progenitors from day 4 EBs (B) but not in Flk-1+ vascular progenitors, which are enriched for the endothelial-specific miRNA, miR-126 (C). (D) qRT-PCR showing expression kinetics of miR-1 and miR-133 during days 4–10 of EB differentiation.
Figure 2. Effects of miR-1 and miR-133 on mesoderm differentiation
(A) Schematic of methods used to express miRNAs in mES cells. mES cells were infected with lentiviruses expressing miR-1 or miR-133 under control of a heterologous EF-1 promoter. Stably infected cells were selected based on their resistance to blasticidin in order to generate stable miRNA-expressing mES cell lines (mES_miR-1_ and mES_miR-133_). (B) qRT-PCR results confirmed the expression of miR-1 and miR-133; expression of the unintroduced miRNA was unchanged. miR-1 and miR-133 were expressed at levels comparable to those in the adult mouse heart. (C) The population doubling times of mES_miR-1_ and mES_miR-133_ cells were similar to those of wild-type mES cells. (D) qRT-PCR analyzing expression of Bry, an early mesoderm marker, in control, mES_miR-1_, and mES_miR-133_ EBs collected on day 4 of differentiation. Expression of miR-1 or miR-133 increased expression of Bry. (E, F) qRT-PCR analysis of Nkx2.5 (E) and Myogenin (F) expression from day 4, 6, or 10 EBs formed from control, mES_miR-1_, or mES_miR-133_ cells. Control EBs displayed an induction of Nkx2.5 expression over time that was enhanced by miR-1 and suppressed by miR-133. Induction of Myogenin expression was enhanced by miR-1, but not by miR-133. (G) Differences in Nkx2.5 expression (green fluorescent cells) were also visualized at day 10 of differentiation by expressing the miRNAs in an _Nkx2.5_-GFP transgenic mES cell line. (H) Expression of miR-1 and miR-133 was undetectable in day 10 _SRF_−/− EBs by qRT-PCR. (I) Overexpression of miR-1 and to a lesser extent, miR-133, in _SRF_−/− EBs restored the Bry and Mesp1 downregulation in day 10 EBs typical of wild-type cells. (J) Expression of Cd53, Cxcl4, and Thbs1, which mark hematopoietic lineages, and of Mef2c, which encodes a major regulator of muscle differentiation, was partially rescued in _SRF_−/− EBs upon expression of miR-1 or miR-133.
Figure 3. Both miR-1 and miR-133 suppress endoderm and neuroectoderm differentiation in mES cells
(A, B) qRT-PCR analysis of the endoderm markers Afp (A) or Hnf4α (B) from day 4, 6, or 10 nodal-treated EBs formed from control, mES_miR-1_ or mES_miR-133_ cells. Induction of Afp and Hnf4α expression normally observed during differentiation in the presence of nodal was suppressed by expression of miR-1 or miR-133. (C) qRT-PCR analysis of the neural marker Ncam1 from day 4, 6, or 10 RA-treated EBs formed from control, mES_miR-1_ or mES_miR-133_ cells. Expression of miR-1 or miR-133 suppressed the induction of Ncam normally observed during differentiation in the presence of RA. (D) qRT-PCR analysis of the neural progenitor marker Nestin in day 4, 8, or 10 RA-treated EBs formed from control, mES_miR-1_ or mES_miR-133_ cells. Nestin expression declined in wild-type EBs by day 10 as neurons differentiated, but was maintained in mES_miR-1_ and mES_miR-133_ EBs. (E) Plot comparing results from mRNA expression microarray analyses of day 10 control, mES_miR-1_, and mES_miR-133_ EBs. Plot shows that most genes were coordinately regulated. (F) Examples of genes that were coordinately regulated in mES_miR-1_ and mES_miR-133_ EBs compared to controls.
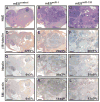
Figure 4. Differentiation of neural cells is suppressed by miR-1 or miR-133 in teratomas
Teratomas were generated by injecting control, mES_miR-1_, or mES_miR-133_ cells into the rear flank of SCID mice. After 6 weeks, hematoxylin/eosin-stained teratomas derived from control ES cells were strikingly homogeneous (A) and composed mostly of βIII-tubulin-immunoreactive neural cells (D). Teratomas from mES_miR-1_ or mES_miR-133_ cells were more heterogeneous (B,C) and contained fewer βIII-tubulin-positive cells (E,F). An accumulation of nestin-positive neural precursors was observed in miR-1 or miR-133 expressing teratomas compared to control (G-I). Expression of miR-1 or miR-133 enhanced muscle specification, as shown by immunostaining with smooth muscle α-actin antibody (J-L). Quantification of areas immunostained with each antibody is indicated as percentages with standard deviation. Scale bars represent ~2mm.
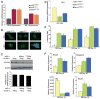
Figure 5. Dll-1 protein levels are negatively regulated by miR-1 in mES cells, and knockdown of Dll-1 expression promotes cardiac mesoderm and suppresses non-mesodermal gene expression
(A) Dll-1 and Dll-4 mRNA levels, assessed by qRT-PCR, were somewhat higher in mES_miR-1_ and mES_miR-133_ cells than in controls. (B) Immunostaining with Dll-4 or Dll-1 antibody showed equivalent Dll-4 protein levels in mES_miR-1_, mES_miR-133_ cells, and control mES cells; Dll-1 protein levels were lower in mES_miR-1_ cells and higher in mES_miR-133_ cells than wild–type mES cells. (C) miR-1 expression caused a dose-dependent decrease in epitope (V5)-tagged Dll-1 protein levels by Western blot without affecting RNA expression of Dll-1 assessed by qRT-PCR (graph). Gapdh protein levels reflect equal loading of protein. (D) Dll-1 mRNA levels, assessed by qRT-PCR, were 62% and 40% lower in response to two distinct short hairpin RNAs targeting Dll-1 mRNA (_Dll-1_shRNA-1 and _Dll-1_shRNA-2), compared to control cell line. (E) EBs formed from _Dll-1_control, _Dll-1_shRNA-1 and _Dll-1_shRNA-2 ES cells were scored for beating cardiomyocytes on days 8, 10, and 12 of differentiation. Beating cardiomyocytes appeared earlier and were more numerous in EBs from _Dll-1_shRNA cell lines than in EBs from the control line. (F) qRT-PCR analyses of Nkx2.5, Myogenin, Afp, and Nestin expression in EBs generated from _Dll-1_control, _Dll-1_shRNA-1 and _Dll-1_shRNA-2 ES cells. Knocking down Dll-1 increased Myogenin expression, decreased Afp expression and sustained Nestin expression compared to controls.
Figure 6. Effects of miR-1 or miR-133 expression in hES cells
(A) Lentivirus-mediated expression of miR-1 or miR-133 in human ES (hES) cells was verified by qRT-PCR. (B) NKX2.5 mRNA expression assessed by qRT-PCR in hEBs collected on days 4, 6, and 8. Overexpression of miR-1 in hES cells increased NKX2.5 expression compared to wild type, while miR-133 expression led to decreased NKX2.5 induction. (C) Human EBs were scored for beating on day 18 of differentiation. Expression of miR-1 increased the number of beating human EBs, while expression of miR-133 did not. (D) Day 18 human EBs were immunostained with antibodies to nestin or βIII-tubulin.
Figure 7. Model of miR-1/miR-133 effects during ES cell differentiation
miR-1 and miR-133 promotion of mesoderm and inhibition of endoderm and ectoderm differentiation at specific stages are indicated. Opposing effects of the two miRNAs in later steps of muscle differentiation are also shown. miR-1 inhibition of Dll-1 translation, along with yet unknown targets, likely contribute to the some of the observed effects of miR-1.
Comment in
- MicroRNA regulation of stem cell fate.
Li Q, Gregory RI. Li Q, et al. Cell Stem Cell. 2008 Mar 6;2(3):195-6. doi: 10.1016/j.stem.2008.02.008. Cell Stem Cell. 2008. PMID: 18371442
Similar articles
- MicroRNA regulation of stem cell fate.
Li Q, Gregory RI. Li Q, et al. Cell Stem Cell. 2008 Mar 6;2(3):195-6. doi: 10.1016/j.stem.2008.02.008. Cell Stem Cell. 2008. PMID: 18371442 - Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Gunaratne PH. Gunaratne PH. Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review. - miRNA-1 and miRNA-133a are involved in early commitment of pluripotent stem cells and demonstrate antagonistic roles in the regulation of cardiac differentiation.
Izarra A, Moscoso I, Cañón S, Carreiro C, Fondevila D, Martín-Caballero J, Blanca V, Valiente I, Díez-Juan A, Bernad A. Izarra A, et al. J Tissue Eng Regen Med. 2017 Mar;11(3):787-799. doi: 10.1002/term.1977. Epub 2014 Dec 10. J Tissue Eng Regen Med. 2017. PMID: 25492026 - Mesoderm cell development from ES cells.
Era T. Era T. Methods Mol Biol. 2010;636:87-103. doi: 10.1007/978-1-60761-691-7_6. Methods Mol Biol. 2010. PMID: 20336518 - The Expression and Functional Roles of miRNAs in Embryonic and Lineage-Specific Stem Cells.
Farzaneh M, Alishahi M, Derakhshan Z, Sarani NH, Attari F, Khoshnam SE. Farzaneh M, et al. Curr Stem Cell Res Ther. 2019;14(3):278-289. doi: 10.2174/1574888X14666190123162402. Curr Stem Cell Res Ther. 2019. PMID: 30674265 Review.
Cited by
- Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth.
Ghanbarian H, Wagner N, Polo B, Baudouy D, Kiani J, Michiels JF, Cuzin F, Rassoulzadegan M, Wagner KD. Ghanbarian H, et al. PLoS One. 2016 Jun 6;11(6):e0156953. doi: 10.1371/journal.pone.0156953. eCollection 2016. PLoS One. 2016. PMID: 27270731 Free PMC article. - Analysis of epigenetic factors in mouse embryonic neural stem cells exposed to hyperglycemia.
Shyamasundar S, Jadhav SP, Bay BH, Tay SS, Kumar SD, Rangasamy D, Dheen ST. Shyamasundar S, et al. PLoS One. 2013 Jun 11;8(6):e65945. doi: 10.1371/journal.pone.0065945. Print 2013. PLoS One. 2013. PMID: 23776576 Free PMC article. - Insights into role of microRNAs in cardiac development, cardiac diseases, and developing novel therapies.
Arabian M, Mirzadeh Azad F, Maleki M, Malakootian M. Arabian M, et al. Iran J Basic Med Sci. 2020 Aug;23(8):961-969. doi: 10.22038/ijbms.2020.40974.10015. Iran J Basic Med Sci. 2020. PMID: 32952941 Free PMC article. Review. - Epigenetics: judge, jury and executioner of stem cell fate.
Tollervey JR, Lunyak VV. Tollervey JR, et al. Epigenetics. 2012 Aug;7(8):823-40. doi: 10.4161/epi.21141. Epub 2012 Jul 18. Epigenetics. 2012. PMID: 22805743 Free PMC article. Review. - A role for RNA post-transcriptional regulation in satellite cell activation.
Farina NH, Hausburg M, Betta ND, Pulliam C, Srivastava D, Cornelison D, Olwin BB. Farina NH, et al. Skelet Muscle. 2012 Oct 9;2(1):21. doi: 10.1186/2044-5040-2-21. Skelet Muscle. 2012. PMID: 23046558 Free PMC article.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
- Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342–357. - PubMed
- Bain G, Ray WJ, Yao M, Gottlieb DI. Retinoic acid promotes neural and represses mesodermal gene expression in mouse embryonic stem cells in culture. Biochem Biophys Res Commun. 1996;223:691–694. - PubMed
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL007544/HL/NHLBI NIH HHS/United States
- R01 HL057181/HL/NHLBI NIH HHS/United States
- C06RR018928/RR/NCRR NIH HHS/United States
- HL085377/HL/NHLBI NIH HHS/United States
- T32 HL007544/HL/NHLBI NIH HHS/United States
- R21 HL085377/HL/NHLBI NIH HHS/United States
- C06 RR018928/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous